| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,309,668 – 14,309,786 |
| Length | 118 |
| Max. P | 0.954578 |

| Location | 14,309,668 – 14,309,786 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.47 |
| Mean single sequence MFE | -16.29 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.25 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14309668 118 + 22407834 UUU-UCCCCAUUAAUUUCUCAUCACUUGCCAUCAACGGCUGUUUAGCCUGAUACGCAACCAUUAAUGAAUCCCAUUAAGAAAAUACAAUCAACAGCUUGUCGAUCAUUUGCUCUUAUCA ...-..........((((.......((((.((((..((((....))))))))..))))...((((((.....))))))))))...........(((.((.....))...)))....... ( -15.00) >DroSec_CAF1 144353 118 + 1 UUU-UUCCCAUUAAUUUUUCAUCACUUGCCAUCAACGGCUACUUAGUCUGAUACGCAACCAUUAAUGAAUUCCAUUAAGAAAAUACAAUCAACGACUUGUCGAACAUUUGCUCUUAUCA ...-...........(((((.....((((.((((..((((....))))))))..))))...((((((.....))))))))))).........(((....)))................. ( -14.60) >DroSim_CAF1 135099 118 + 1 UUU-UUCCCAUUAAUUUUUCAUCACUUGCCAUCAACGGCUACUUAGCCUGAUACGCAACCAUUAAUGAAUUCCAUUAAGAAAAUACAAUCAACGGCUUGUCGAACAUUUGCUCUUAUCA ...-.................((((..(((......((((....))))((((.........((((((.....)))))).........))))..)))..)).))................ ( -17.97) >DroEre_CAF1 143831 118 + 1 UUU-UUCCCAUUAAUUUCACAUCAGUUCCCAUCAACGGCUGCCGAGCCUGAUACGCAACCAUUAAUGAAUCCCAUUAAGAAAAUACUAUCAACGGCUUGUCGAACAUUUCUUCUUAUCA ...-....................((((((......))..(.(((((((((((........((((((.....))))))........)))))..)))))).))))).............. ( -19.29) >DroYak_CAF1 133393 119 + 1 UUUUUUUUCAUUUAUUUCUCAUCACUUUCCAUCAACGGCUGCUUAGCCUGAUACGCAAACAUUAAUGAAUCCCAUUAAGAAAAUACAAUCAACGGCUUGUCGAACAUUUCUUCUUAUCA ...................................((((.....((((((((.........((((((.....)))))).........))))..)))).))))................. ( -14.57) >consensus UUU_UUCCCAUUAAUUUCUCAUCACUUGCCAUCAACGGCUGCUUAGCCUGAUACGCAACCAUUAAUGAAUCCCAUUAAGAAAAUACAAUCAACGGCUUGUCGAACAUUUGCUCUUAUCA ...................................((((.....((((((((.........((((((.....)))))).........))))..)))).))))................. (-13.37 = -13.25 + -0.12)


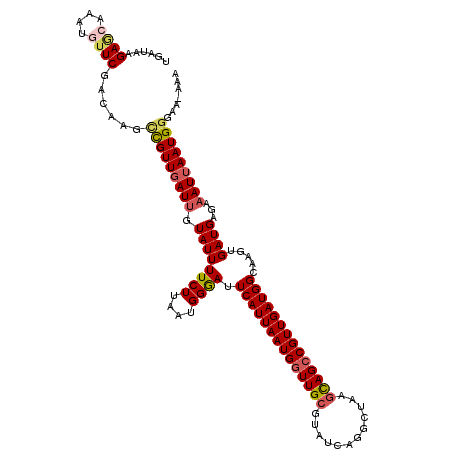
| Location | 14,309,668 – 14,309,786 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.47 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -22.92 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14309668 118 - 22407834 UGAUAAGAGCAAAUGAUCGACAAGCUGUUGAUUGUAUUUUCUUAAUGGGAUUCAUUAAUGGUUGCGUAUCAGGCUAAACAGCCGUUGAUGGCAAGUGAUGAGAAAUUAAUGGGGA-AAA ............(..((((((.....))))))..).((((((((.(((..(((((((....((((.(((((((((....))))..))))))))).)))))))...))).))))))-)). ( -30.00) >DroSec_CAF1 144353 118 - 1 UGAUAAGAGCAAAUGUUCGACAAGUCGUUGAUUGUAUUUUCUUAAUGGAAUUCAUUAAUGGUUGCGUAUCAGACUAAGUAGCCGUUGAUGGCAAGUGAUGAAAAAUUAAUGGGAA-AAA ......((((....))))......(((((((((.((((((((....)))).((((((((((((((............)))))))))))))).....))))...)))))))))...-... ( -27.50) >DroSim_CAF1 135099 118 - 1 UGAUAAGAGCAAAUGUUCGACAAGCCGUUGAUUGUAUUUUCUUAAUGGAAUUCAUUAAUGGUUGCGUAUCAGGCUAAGUAGCCGUUGAUGGCAAGUGAUGAAAAAUUAAUGGGAA-AAA ......((((....))))......(((((((((.....((((....))))(((((((....((((.(((((((((....))))..))))))))).))))))).)))))))))...-... ( -30.70) >DroEre_CAF1 143831 118 - 1 UGAUAAGAAGAAAUGUUCGACAAGCCGUUGAUAGUAUUUUCUUAAUGGGAUUCAUUAAUGGUUGCGUAUCAGGCUCGGCAGCCGUUGAUGGGAACUGAUGUGAAAUUAAUGGGAA-AAA ...((((((((.((..(((((.....)))))..)).))))))))..((..(((((((((((((((............)))))))))))))))..))...................-... ( -31.00) >DroYak_CAF1 133393 119 - 1 UGAUAAGAAGAAAUGUUCGACAAGCCGUUGAUUGUAUUUUCUUAAUGGGAUUCAUUAAUGUUUGCGUAUCAGGCUAAGCAGCCGUUGAUGGAAAGUGAUGAGAAAUAAAUGAAAAAAAA ...((((((((.((..(((((.....)))))..)).))))))))......(((((((...(((.(((....((((....))))....))).))).)))))))................. ( -25.50) >consensus UGAUAAGAGCAAAUGUUCGACAAGCCGUUGAUUGUAUUUUCUUAAUGGGAUUCAUUAAUGGUUGCGUAUCAGGCUAAGCAGCCGUUGAUGGCAAGUGAUGAGAAAUUAAUGGGAA_AAA ......((((....))))......(((((((((.((((((((....)))).((((((((((((((............)))))))))))))).....))))...)))))))))....... (-22.92 = -23.60 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:16 2006