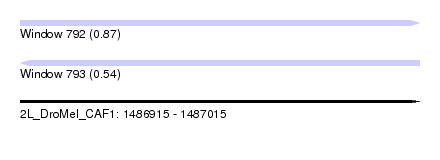
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,486,915 – 1,487,015 |
| Length | 100 |
| Max. P | 0.868350 |

| Location | 1,486,915 – 1,487,015 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.45 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -18.73 |
| Energy contribution | -18.12 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
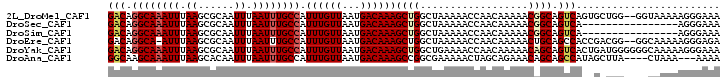
>2L_DroMel_CAF1 1486915 100 + 22407834 UUUCCCUUUUUACC--CCAGCACUGACUGCCGUUUUUGUUGGUUUUUAGCCAGCUUUGUCAUUAACAAAUGGCAAAUUAAAUUGCGCUUAAAUUUGCCUGUC ..............--.(((((.(((.(((.((((..((((((.....))))))(((((((((....)))))))))..)))).))).)))....)).))).. ( -19.10) >DroSec_CAF1 104731 86 + 1 UUUCCCU----------------UGACUGCCGUUUUUGUUGGUUUUUAGCCAGCUUUGUCAUUAACAAAUGGCAAAUUAAAUUGCGCUUAAAUUUGCCUGUC .......----------------.(((.((.((((..((((((.....))))))(((((((((....)))))))))..)))).))((........))..))) ( -18.80) >DroSim_CAF1 107025 86 + 1 UUUCCCU----------------UGACUGCCGUUUUUGUUGGUUUUUAGCCAGCUUUGUCAUUAACAAAUGGCAAAUUAAAUUGCGCUUAAAUUUGCCUGUC .......----------------.(((.((.((((..((((((.....))))))(((((((((....)))))))))..)))).))((........))..))) ( -18.80) >DroEre_CAF1 110563 99 + 1 UCUCCCUUUUUGCC--CCGUCGGUGGCUGCAGUUUUUGUUGGUUUUUAGCCAGCUUUGUCAUUAACAAAUGGCAAAUUAAAUUGCGCUUAAAU-UGCCUGUC ...........(((--((...)).))).(((((((..((((((.....))))))(((((((((....)))))))))..)))))))((......-.))..... ( -26.10) >DroYak_CAF1 107604 102 + 1 UUUCCCUUUUUGCCCCCCAUCAGUGACUGCUGUUUUUGUUGGUUUUCAGCCAGCUUUGUCAUUAACAAAUGGCAAAUUAAAUUGCGCUUAAAUUUGCCUGUC .....................((((((.((((.....((((.....))))))))...)))))).......((((((((.((......)).)))))))).... ( -20.20) >DroAna_CAF1 106058 95 + 1 UUUU---UUUAG----UAAGCUAUGGCUGCUGUUUCUGCUAGUUUUUCGCCGGCUUUGUCAUUAACAAAUGGCAAAUUAAAUUGUGCUUAAAUUUGCUUGCC ....---....(----(((((...(((.((.((((..((..((.....))..))(((((((((....)))))))))..)))).))))).......)))))). ( -20.20) >consensus UUUCCCUUUUUG____CCA_C__UGACUGCCGUUUUUGUUGGUUUUUAGCCAGCUUUGUCAUUAACAAAUGGCAAAUUAAAUUGCGCUUAAAUUUGCCUGUC ........................(((.((.((((..((((((.....))))))(((((((((....)))))))))..)))).))((........))..))) (-18.73 = -18.12 + -0.61)

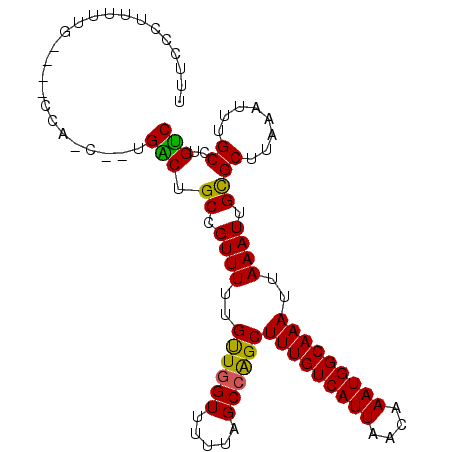
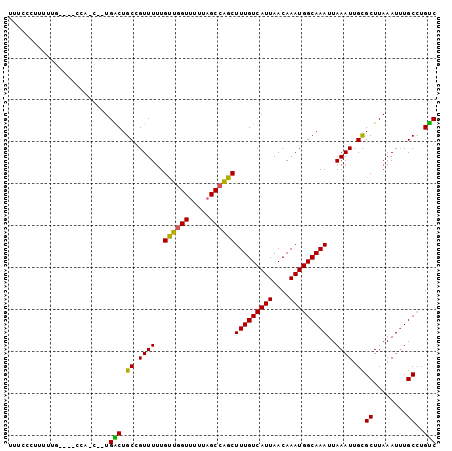
| Location | 1,486,915 – 1,487,015 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.45 |
| Mean single sequence MFE | -18.65 |
| Consensus MFE | -15.12 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1486915 100 - 22407834 GACAGGCAAAUUUAAGCGCAAUUUAAUUUGCCAUUUGUUAAUGACAAAGCUGGCUAAAAACCAACAAAAACGGCAGUCAGUGCUGG--GGUAAAAAGGGAAA ((((((((((((.((......)).))))))))...)))).((..((..(((((((.....((.........)).)))))))..)).--.))........... ( -18.90) >DroSec_CAF1 104731 86 - 1 GACAGGCAAAUUUAAGCGCAAUUUAAUUUGCCAUUUGUUAAUGACAAAGCUGGCUAAAAACCAACAAAAACGGCAGUCA----------------AGGGAAA (((.((((((((.((......)).)))))))).((((((...))))))((((..................)))).))).----------------....... ( -18.37) >DroSim_CAF1 107025 86 - 1 GACAGGCAAAUUUAAGCGCAAUUUAAUUUGCCAUUUGUUAAUGACAAAGCUGGCUAAAAACCAACAAAAACGGCAGUCA----------------AGGGAAA (((.((((((((.((......)).)))))))).((((((...))))))((((..................)))).))).----------------....... ( -18.37) >DroEre_CAF1 110563 99 - 1 GACAGGCA-AUUUAAGCGCAAUUUAAUUUGCCAUUUGUUAAUGACAAAGCUGGCUAAAAACCAACAAAAACUGCAGCCACCGACGG--GGCAAAAAGGGAGA ....((((-(.(((((.....))))).))))).((((((...))))))(((((((...................)))))((....)--)))........... ( -17.51) >DroYak_CAF1 107604 102 - 1 GACAGGCAAAUUUAAGCGCAAUUUAAUUUGCCAUUUGUUAAUGACAAAGCUGGCUGAAAACCAACAAAAACAGCAGUCACUGAUGGGGGGCAAAAAGGGAAA .....((........)).........((((((.((..(((.((((...((((..................)))).)))).)))..)).))))))........ ( -21.57) >DroAna_CAF1 106058 95 - 1 GGCAAGCAAAUUUAAGCACAAUUUAAUUUGCCAUUUGUUAAUGACAAAGCCGGCGAAAAACUAGCAGAAACAGCAGCCAUAGCUUA----CUAAA---AAAA (((..((........)).........((((((.((((((...))))))...))))))......((.......)).)))........----.....---.... ( -17.20) >consensus GACAGGCAAAUUUAAGCGCAAUUUAAUUUGCCAUUUGUUAAUGACAAAGCUGGCUAAAAACCAACAAAAACGGCAGUCA__G_UGG____CAAAAAGGGAAA (((.((((((((.((......)).)))))))).((((((...))))))((((..................)))).)))........................ (-15.12 = -15.65 + 0.53)
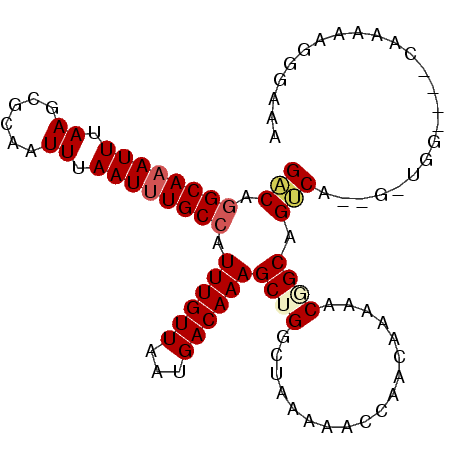
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:33 2006