| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,304,038 – 14,304,137 |
| Length | 99 |
| Max. P | 0.528914 |

| Location | 14,304,038 – 14,304,137 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.48 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -10.68 |
| Energy contribution | -11.18 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
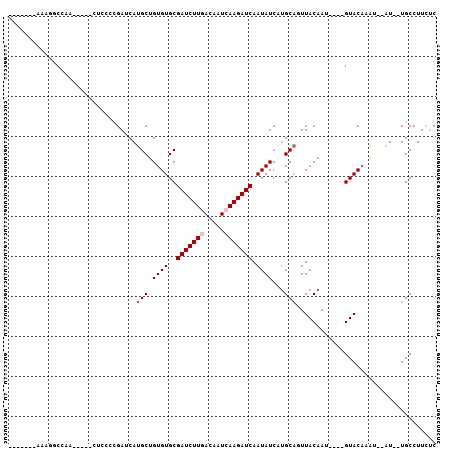
>2L_DroMel_CAF1 14304038 99 + 22407834 UAGUCACAAAGGCCAACUCUCCCCUCCGAUCAUGCUGUGUGCGAUCUUAACAAUCAAGAUCAAUAUCAUGCAUUUACAAU----GUACAAAU--AU--UGCCUUCUC ........(((((..................(((((((((..((((((.......)))))).))).)).))))....(((----((....))--))--))))))... ( -15.20) >DroPse_CAF1 168959 87 + 1 -------AAGAGCUGG-----CUCCCCGAGCAUGCUGUGUGCGAUCUUGACAAUCAAGAUCAAUAUCUUGCAGUUACAAU----GUACAAAU--AU--UGCCUUCUC -------..(((..((-----(.......((((.....))))(((((((.....)))))))(((((..((((.......)----)))...))--))--))))..))) ( -22.70) >DroGri_CAF1 149644 95 + 1 ---------CGACGAA-CGUAGUCUGCGAUCAUGCUGUGUGCGAUCUUGACAAUCAAGAUCAAUAUCAUGCCAUUACAAUAUUUGUACAAGACGCU--UGCCUUUAC ---------.(((...-....))).(((.((..(((((((..(((((((.....))))))).))).)).))...((((.....))))...))))).--......... ( -19.50) >DroWil_CAF1 203010 89 + 1 -GACGACAAAGACGA-----------UGAUCAUGCUGUGUGCGAUCUUGACAAUCAAGAUCAAUAUCAGGCAUUUACAAU----GUACAAAU--AUUUUGCCUUCUC -............((-----------((.....((.....))(((((((.....)))))))..))))(((((.....(((----((....))--))).))))).... ( -18.90) >DroAna_CAF1 141202 84 + 1 ------------CCAACUC---CCCUCGAUCAUGCUGUGUGCGAUCUUAACAAUCAAGAUCAAUAUCAUGCAGUUACAAU----GUACCAAU--AU--UGCCUUCUC ------------.......---...........((((((((.((((((.......)))))).....))))))))..((((----((....))--))--))....... ( -13.80) >DroPer_CAF1 174570 87 + 1 -------AAGAGCUGG-----CUCCCCGAGCAUGCUGUGUGCGAUCUUGACAAUCAAGAUCAAUAUCUUGCAGUUACAAU----GUACAAAU--AU--UGCCUUCUC -------..(((..((-----(.......((((.....))))(((((((.....)))))))(((((..((((.......)----)))...))--))--))))..))) ( -22.70) >consensus _______AAAGGCCAA_____CUCCCCGAUCAUGCUGUGUGCGAUCUUGACAAUCAAGAUCAAUAUCAUGCAGUUACAAU____GUACAAAU__AU__UGCCUUCUC ................................(((.((((..(((((((.....))))))).))))...)))................................... (-10.68 = -11.18 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:13 2006