| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,272,057 – 14,272,171 |
| Length | 114 |
| Max. P | 0.822749 |

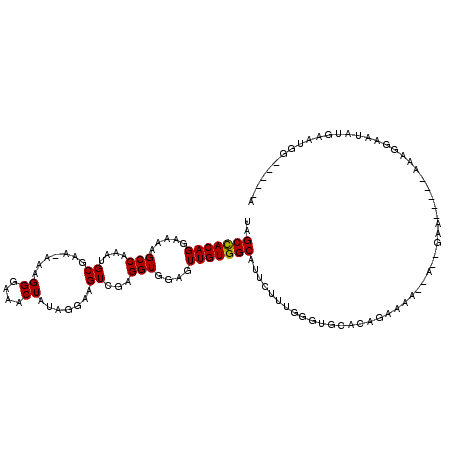
| Location | 14,272,057 – 14,272,171 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.14 |
| Mean single sequence MFE | -13.46 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14272057 114 + 22407834 UCCAUUCCAUUCAUAUUCCUUUUUU--UUC--U--UUUUCUGUGCACCCAAAGAAUGCCACAACUCCACCUCGACUUCCUAUAGUUUCCCUUUUUUUGCAUUUGGCUUUUCCUGUGGCUA .........................--...--.--..((((.((....)).)))).((((((..........((((......))))...........((.....))......)))))).. ( -12.60) >DroSec_CAF1 104692 101 + 1 U-----CCAUUCAUAUUCCUU------UUC--U--UUUUCUGUGCACCCAA---AUGCCACAACUCCACCUCGACUUCCUAUAGUUUCCCUUU-UUCGCAUUUGGCUUUUCCUGUGGCUA .-----...............------...--.--................---..((((((..........((((......)))).......-...((.....))......)))))).. ( -11.70) >DroSim_CAF1 96774 104 + 1 U-----CCAUUCAUAUUCCUU------UUC--U--UUUUCUGUGCACCCAAAGCAUGCCACAACUCCACCUCGACUUCCUAUAGUUUCCCUUU-UUCGCAUUUGGCUUUUCCUGUGGCUA .-----...............------...--.--......((((.......))))((((((..........((((......)))).......-...((.....))......)))))).. ( -14.60) >DroEre_CAF1 105915 114 + 1 U-----CCGUUCAUACUUCAUUAUUAUCUCGCUUCUUUGUUGUGCCACCAAAGAAUGCCACAACUCCACCUCAACUUCCUAAAGUUUCCCUUU-UUCGCAUUUGGCUUUUCCUGUAGCCA .-----........................((......((((((.((........)).))))))........(((((....))))).......-...))...(((((........))))) ( -15.60) >DroYak_CAF1 95535 111 + 1 U-----CCGUUCGUACUUUAAUCCCAUCUCGUUUCUUU--CGUGCACCCAAAGAAUGCCACAACUCCACCUCGACUUCCUCUAGUUUCCCUUU-UU-GCAUUUGGCUUUUCCUGUGGCCA .-----..........................((((((--.(.....).)))))).((((((..........((((......)))).......-..-((.....))......)))))).. ( -12.80) >consensus U_____CCAUUCAUAUUCCUUU_____UUC__U__UUUUCUGUGCACCCAAAGAAUGCCACAACUCCACCUCGACUUCCUAUAGUUUCCCUUU_UUCGCAUUUGGCUUUUCCUGUGGCUA ........................................................((((((..........((((......))))...........((.....))......)))))).. (-10.12 = -10.16 + 0.04)

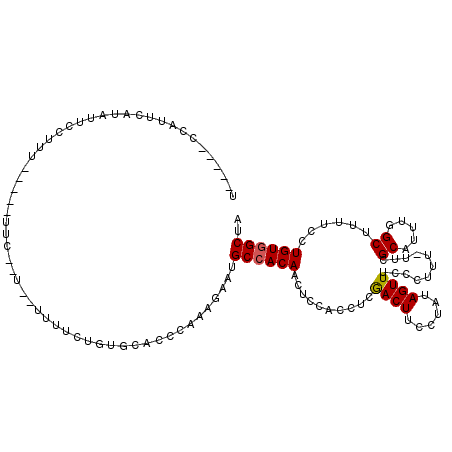
| Location | 14,272,057 – 14,272,171 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.14 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -16.06 |
| Energy contribution | -15.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14272057 114 - 22407834 UAGCCACAGGAAAAGCCAAAUGCAAAAAAAGGGAAACUAUAGGAAGUCGAGGUGGAGUUGUGGCAUUCUUUGGGUGCACAGAAAA--A--GAA--AAAAAAGGAAUAUGAAUGGAAUGGA ..(((((((.....(((.............((....))....(....)..)))....))))))).((((((.(.....)....))--)--)))--......................... ( -18.90) >DroSec_CAF1 104692 101 - 1 UAGCCACAGGAAAAGCCAAAUGCGAA-AAAGGGAAACUAUAGGAAGUCGAGGUGGAGUUGUGGCAU---UUGGGUGCACAGAAAA--A--GAA------AAGGAAUAUGAAUGG-----A ..(((((((......(((....(((.-...((....))........)))...)))..))))))).(---(((......))))...--.--...------...............-----. ( -19.20) >DroSim_CAF1 96774 104 - 1 UAGCCACAGGAAAAGCCAAAUGCGAA-AAAGGGAAACUAUAGGAAGUCGAGGUGGAGUUGUGGCAUGCUUUGGGUGCACAGAAAA--A--GAA------AAGGAAUAUGAAUGG-----A ..(((((((......(((....(((.-...((....))........)))...)))..))))))).(((.......))).......--.--...------...............-----. ( -19.50) >DroEre_CAF1 105915 114 - 1 UGGCUACAGGAAAAGCCAAAUGCGAA-AAAGGGAAACUUUAGGAAGUUGAGGUGGAGUUGUGGCAUUCUUUGGUGGCACAACAAAGAAGCGAGAUAAUAAUGAAGUAUGAACGG-----A (((((........)))))..(((...-.((((....))))................((((((.(((......))).))))))......))).......................-----. ( -24.90) >DroYak_CAF1 95535 111 - 1 UGGCCACAGGAAAAGCCAAAUGC-AA-AAAGGGAAACUAGAGGAAGUCGAGGUGGAGUUGUGGCAUUCUUUGGGUGCACG--AAAGAAACGAGAUGGGAUUAAAGUACGAACGG-----A ..(((((((.....(((......-..-...((....)).((.....))..)))....))))))).((((((.(.....).--))))))..........................-----. ( -22.20) >consensus UAGCCACAGGAAAAGCCAAAUGCGAA_AAAGGGAAACUAUAGGAAGUCGAGGUGGAGUUGUGGCAUUCUUUGGGUGCACAGAAAA__A__GAA_____AAAGGAAUAUGAAUGG_____A ..(((((((.....(((....((.......((....)).......))...)))....)))))))........................................................ (-16.06 = -15.90 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:12 2006