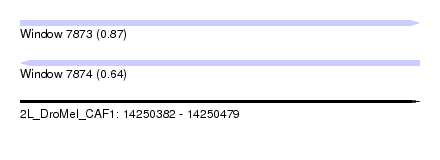
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,250,382 – 14,250,479 |
| Length | 97 |
| Max. P | 0.874967 |

| Location | 14,250,382 – 14,250,479 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -18.49 |
| Consensus MFE | -13.65 |
| Energy contribution | -13.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14250382 97 + 22407834 GAAACUGACAGUUUGACGUUUUAACUCACGCGCCAAACG-------CCAAACGUCAAACGGCAGCAACAACCAACA--CAAUAUCAACAUC------AGCAACAUAGUAAUA ....((((..(((((((((((........(((.....))-------).)))))))))))((.........))....--...........))------))............. ( -17.40) >DroSec_CAF1 83228 90 + 1 GAAACUGACAGUUUGACGUUUUAACUCACGCGCCA--------------AACGUCAAACGGCAGCAACAACCAACA--CAAUAUCAACAUC------AGCAACAUAGUAAUA ....((((..(((((((((((.............)--------------))))))))))((.........))....--...........))------))............. ( -15.52) >DroSim_CAF1 75210 90 + 1 GAAACUGACAGUUUGACGUUUUAACUCACGCGCCA--------------AACGUCAAACGGCAGCAACAACCAACA--CAAUAUCAACAUC------AGCAACAUAGUAAUA ....((((..(((((((((((.............)--------------))))))))))((.........))....--...........))------))............. ( -15.52) >DroEre_CAF1 84131 106 + 1 GAAACUGACAGUUUGACGUUUUAACUCACGCGCCAAACGGCAAACGCCAAACGUCAAACGGCAGCAACAACCAACACACAAUAUCAACAUC------AGCAACAUAGUAAUA ....((((..(((((((((((..........(.....)(((....))))))))))))))((.........)).................))------))............. ( -19.60) >DroYak_CAF1 72636 104 + 1 GAAGCUGACAGUUUGACGUUUUAACUCACGCGCCAAACGCCAAACGCUAAACGUCAAACGGCAGCAACAACCAACA--CAAUAUCAACAUC------AGCAACAUAGUAAUA ...(((((..(((((((((((........(((.....)))........)))))))))))((.........))....--...........))------)))............ ( -24.89) >DroAna_CAF1 85372 92 + 1 GGAACUGACAGUUUGACGUUUUAACUCACGCGCCA--------------AACGUCAAACGGCAGCUACGA----AA--CAAUGGCCAUACCCCAAUACCCCAUACCCCAAUA ((..(((...(((((((((((.............)--------------))))))))))..)))......----..--...(((.......))).....))........... ( -18.02) >consensus GAAACUGACAGUUUGACGUUUUAACUCACGCGCCA______________AACGUCAAACGGCAGCAACAACCAACA__CAAUAUCAACAUC______AGCAACAUAGUAAUA ....(((...((((((((((.............................))))))))))..)))................................................ (-13.65 = -13.65 + 0.00)

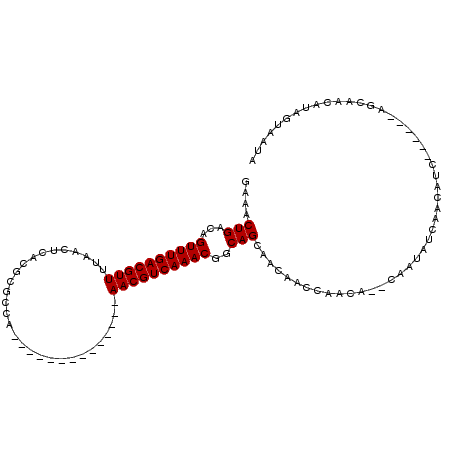
| Location | 14,250,382 – 14,250,479 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.05 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
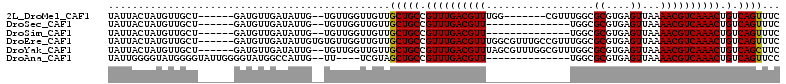
>2L_DroMel_CAF1 14250382 97 - 22407834 UAUUACUAUGUUGCU------GAUGUUGAUAUUG--UGUUGGUUGUUGCUGCCGUUUGACGUUUGG-------CGUUUGGCGCGUGAGUUAAAACGUCAAACUGUCAGUUUC ............(((------(((((..(.....--...((((.......)))).)..))((((((-------(((((...((....))..))))))))))).))))))... ( -27.10) >DroSec_CAF1 83228 90 - 1 UAUUACUAUGUUGCU------GAUGUUGAUAUUG--UGUUGGUUGUUGCUGCCGUUUGACGUU--------------UGGCGCGUGAGUUAAAACGUCAAACUGUCAGUUUC ............(((------(((..........--....(((.......)))((((((((((--------------(...((....))..))))))))))).))))))... ( -26.60) >DroSim_CAF1 75210 90 - 1 UAUUACUAUGUUGCU------GAUGUUGAUAUUG--UGUUGGUUGUUGCUGCCGUUUGACGUU--------------UGGCGCGUGAGUUAAAACGUCAAACUGUCAGUUUC ............(((------(((..........--....(((.......)))((((((((((--------------(...((....))..))))))))))).))))))... ( -26.60) >DroEre_CAF1 84131 106 - 1 UAUUACUAUGUUGCU------GAUGUUGAUAUUGUGUGUUGGUUGUUGCUGCCGUUUGACGUUUGGCGUUUGCCGUUUGGCGCGUGAGUUAAAACGUCAAACUGUCAGUUUC ............(((------(((((..(..........((((.......)))).)..))((((((((((((((....)))((....))..))))))))))).))))))... ( -29.20) >DroYak_CAF1 72636 104 - 1 UAUUACUAUGUUGCU------GAUGUUGAUAUUG--UGUUGGUUGUUGCUGCCGUUUGACGUUUAGCGUUUGGCGUUUGGCGCGUGAGUUAAAACGUCAAACUGUCAGCUUC ............(((------((((((((...((--(..((((.......))))....))).)))))(((((((((((...((....))..))))))))))).))))))... ( -32.30) >DroAna_CAF1 85372 92 - 1 UAUUGGGGUAUGGGGUAUUGGGGUAUGGCCAUUG--UU----UCGUAGCUGCCGUUUGACGUU--------------UGGCGCGUGAGUUAAAACGUCAAACUGUCAGUUCC ....(((.(((((..((.(((.......))).))--..----)))))(((((.((((((((((--------------(...((....))..))))))))))).).))))))) ( -31.00) >consensus UAUUACUAUGUUGCU______GAUGUUGAUAUUG__UGUUGGUUGUUGCUGCCGUUUGACGUU______________UGGCGCGUGAGUUAAAACGUCAAACUGUCAGUUUC ...............................................(((((.((((((((((..................((....))...)))))))))).).))))... (-17.19 = -17.05 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:07 2006