
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
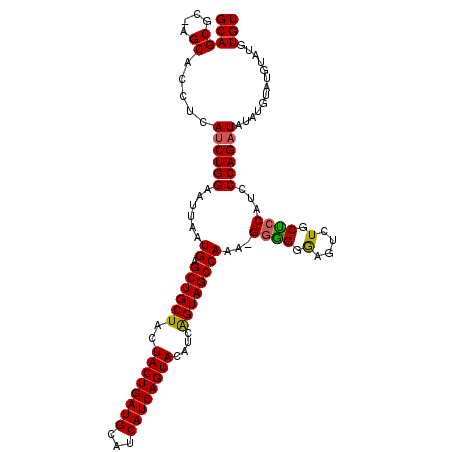
| Location | 14,240,229 – 14,240,372 |
| Length | 143 |
| Max. P | 0.910806 |

| Location | 14,240,229 – 14,240,341 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -29.48 |
| Energy contribution | -29.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14240229 112 + 22407834 GCAGCGCCAGCACCUCAUCUGGAAUUAAUGAGCUGCUCCUACUGAUGCAUCAUCAGUACAUCGGUAGCCAAA-GGGGGAAGUCUGCCACAUCCCAGAUAAA-GUAUGUAUAUGU (((((....)).....((((((......((.((((((..((((((((...))))))))....))))))))..-((.((....)).)).....))))))...-...)))...... ( -32.30) >DroSec_CAF1 72923 113 + 1 GCAGCGC-AGCACCUCAUCUGGAAUUAAUGAGCUGCUACUACUGAUGCAUCAUCAGUACAUCAGUAGCCAAACGGAGGGAGUCUGCUCCAUCCCAGCUAUAUGUAUGUAUGUGU ...((((-((((...(((.(((......((.((((((..((((((((...))))))))....))))))))...(((.((((....)))).)))...))).)))..))).))))) ( -35.30) >DroSim_CAF1 65157 108 + 1 GCAGCGC-AGCACCUCAUCUGGAAUUAAUGAGCUGCUACUACUGAUGCAUCAUCAGUACAUCAGUAGCCAAA-GGGGGGAGUCUGCUCCAUCCCAGAUAUAU----GUAUGUGU ...((((-((((....((((........((.((((((..((((((((...))))))))....))))))))..-((((((((....)))).))))))))...)----)).))))) ( -36.30) >consensus GCAGCGC_AGCACCUCAUCUGGAAUUAAUGAGCUGCUACUACUGAUGCAUCAUCAGUACAUCAGUAGCCAAA_GGGGGGAGUCUGCUCCAUCCCAGAUAUAUGUAUGUAUGUGU (((((....)).....((((((......((.((((((..((((((((...))))))))....))))))))...((((.(....).))))...)))))).............))) (-29.48 = -29.27 + -0.22)


| Location | 14,240,229 – 14,240,341 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -39.83 |
| Consensus MFE | -32.32 |
| Energy contribution | -33.10 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14240229 112 - 22407834 ACAUAUACAUAC-UUUAUCUGGGAUGUGGCAGACUUCCCCC-UUUGGCUACCGAUGUACUGAUGAUGCAUCAGUAGGAGCAGCUCAUUAAUUCCAGAUGAGGUGCUGGCGCUGC .......(((((-(((((((((((((((((.(.((((....-.((((...))))..((((((((...))))))))))))).)).)))..))))))))))))))).))....... ( -35.70) >DroSec_CAF1 72923 113 - 1 ACACAUACAUACAUAUAGCUGGGAUGGAGCAGACUCCCUCCGUUUGGCUACUGAUGUACUGAUGAUGCAUCAGUAGUAGCAGCUCAUUAAUUCCAGAUGAGGUGCU-GCGCUGC ...............((((..(((.((((....)))).))).......(((((((((((....).))))))))))((((((.((((((.......)))))).))))-)))))). ( -42.50) >DroSim_CAF1 65157 108 - 1 ACACAUAC----AUAUAUCUGGGAUGGAGCAGACUCCCCCC-UUUGGCUACUGAUGUACUGAUGAUGCAUCAGUAGUAGCAGCUCAUUAAUUCCAGAUGAGGUGCU-GCGCUGC ........----........(((..((((....)))).)))-...((((((((((((((....).))))))))))((((((.((((((.......)))))).))))-))))).. ( -41.30) >consensus ACACAUACAUACAUAUAUCUGGGAUGGAGCAGACUCCCCCC_UUUGGCUACUGAUGUACUGAUGAUGCAUCAGUAGUAGCAGCUCAUUAAUUCCAGAUGAGGUGCU_GCGCUGC ...............((((((((((.((((......((.......))((((((((((((....).))))))))))).....))))....)))))))))).((((....)))).. (-32.32 = -33.10 + 0.78)



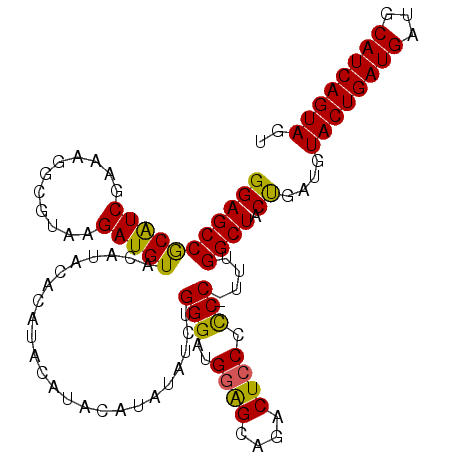
| Location | 14,240,266 – 14,240,372 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14240266 106 + 22407834 CCUACUGAUGCAUCAUCAGUACAUCGGUAGCCAAA-GGGGGAAGUCUGCCACAUCCCAGAUAAA-GUAUGUAUAUGUAUGUACAUCUUACGCCUUUCGACGUGGCUCC ..((((((((...))))))))....((.(((((..-(..((..(((((........))))).((-(.(((((((....))))))))))....))..)....))))))) ( -31.20) >DroSec_CAF1 72959 108 + 1 ACUACUGAUGCAUCAUCAGUACAUCAGUAGCCAAACGGAGGGAGUCUGCUCCAUCCCAGCUAUAUGUAUGUAUGUGUAUGUACAUCUUACGCCUUUCGAUGCGGCUCC .(((((((((.((.....)).)))))))))......(((.((((....)))).))).(((...(((((((((....)))))))))....(((........)))))).. ( -31.40) >DroSim_CAF1 65193 103 + 1 ACUACUGAUGCAUCAUCAGUACAUCAGUAGCCAAA-GGGGGGAGUCUGCUCCAUCCCAGAUAUAU----GUAUGUGUAUGUACAUCUUACGCCUUUCGAUGCGGCUCC .(((((((((.((.....)).))))))))).....-((((((((....)))).))))((((.(((----(((....)))))).))))...(((.........)))... ( -31.00) >consensus ACUACUGAUGCAUCAUCAGUACAUCAGUAGCCAAA_GGGGGGAGUCUGCUCCAUCCCAGAUAUAUGUAUGUAUGUGUAUGUACAUCUUACGCCUUUCGAUGCGGCUCC .(((((((((.((.....)).)))))))))......((((((((....)))).))))............(((.((((....))))..)))(((.........)))... (-25.04 = -25.60 + 0.56)


| Location | 14,240,266 – 14,240,372 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -26.03 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14240266 106 - 22407834 GGAGCCACGUCGAAAGGCGUAAGAUGUACAUACAUAUACAUAC-UUUAUCUGGGAUGUGGCAGACUUCCCCC-UUUGGCUACCGAUGUACUGAUGAUGCAUCAGUAGG (((((((((((....)))))((((((((........))))).)-)).....((((.((.....)).))))..-...)))).))....((((((((...)))))))).. ( -34.50) >DroSec_CAF1 72959 108 - 1 GGAGCCGCAUCGAAAGGCGUAAGAUGUACAUACACAUACAUACAUAUAGCUGGGAUGGAGCAGACUCCCUCCGUUUGGCUACUGAUGUACUGAUGAUGCAUCAGUAGU ((((((((.((....)).(((.(((((......)))).).))).....)).))...((((....)))))))).....(((((((((((((....).)))))))))))) ( -34.60) >DroSim_CAF1 65193 103 - 1 GGAGCCGCAUCGAAAGGCGUAAGAUGUACAUACACAUAC----AUAUAUCUGGGAUGGAGCAGACUCCCCCC-UUUGGCUACUGAUGUACUGAUGAUGCAUCAGUAGU ...(((.........)))...(((((((..((....)).----.)))))))(((..((((....)))).)))-....(((((((((((((....).)))))))))))) ( -32.70) >consensus GGAGCCGCAUCGAAAGGCGUAAGAUGUACAUACACAUACAUACAUAUAUCUGGGAUGGAGCAGACUCCCCCC_UUUGGCUACUGAUGUACUGAUGAUGCAUCAGUAGU (((((((((((...........)))))........................(((..((((....)))).)))....)))).))....((((((((...)))))))).. (-26.03 = -25.27 + -0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:04 2006