| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,237,596 – 14,237,700 |
| Length | 104 |
| Max. P | 0.994069 |

| Location | 14,237,596 – 14,237,700 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.17 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -14.26 |
| Energy contribution | -15.32 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14237596 104 + 22407834 UCUGGAAUGCCCAAUAACCAAGUACUUGAUUAUAGGGGUAAACAAUUUACUAGCUAAUUGGAUUUAAUAAGUGUGCUUAUAUUGCUUAUAUGUGGAAUA------------AAUUA---- .......(((((.((((.(((....))).))))..)))))..(((((........)))))(((((((((((....))))).(..(......)..)..))------------)))).---- ( -16.80) >DroSec_CAF1 70284 98 + 1 --UGGAAUGCCCUAUACCCAAGUACUUGAUUAUAGGGGUA-ACAAUUUAUUAGGUAAUUGAAUUUAAUAAGUGUGUAAAUAUU-----UAUG-GGAAUA------------AAUAAAU-A --.......(((((((..(((....)))..)))))))...-((((((((((((((......))))))))))).)))...((((-----(((.-...)))------------))))...-. ( -21.00) >DroSim_CAF1 62534 98 + 1 --UGGAAUGCCCUAUACCCAAGUACUUGAUUAUAGGGGUA-ACAAUUUAUUAGCUAUUUGAGUUUAAUAAGUGUGUAAAUAUU-----UAUG-GGAAUA------------AAUAAAU-A --.......(((((((..(((....)))..)))))))...-((((((((((((((.....)).))))))))).)))...((((-----(((.-...)))------------))))...-. ( -19.80) >DroEre_CAF1 71293 107 + 1 ------AUGCCCCAUAGCCAAGUACUUGAUCAUAGGGGUA-ACUAUUAAUUAGCUAUUAUUAGCUAAUUAGUGUGUCUUUAUU-----UCUA-UGUCUUACUUUCCAUCUUAAGAAACCG ------..........(..(((((...((.(((((..(((-(((((((((((((((....))))))))))))).))...))).-----.)))-)))).)))))..).............. ( -26.60) >consensus __UGGAAUGCCCUAUACCCAAGUACUUGAUUAUAGGGGUA_ACAAUUUAUUAGCUAAUUGAAUUUAAUAAGUGUGUAAAUAUU_____UAUG_GGAAUA____________AAUAAAU_A .........((((((((.(((....))).))))))))....((.(((((((((((......))))))))))))).............................................. (-14.26 = -15.32 + 1.06)


| Location | 14,237,596 – 14,237,700 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.17 |
| Mean single sequence MFE | -17.34 |
| Consensus MFE | -11.00 |
| Energy contribution | -10.38 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
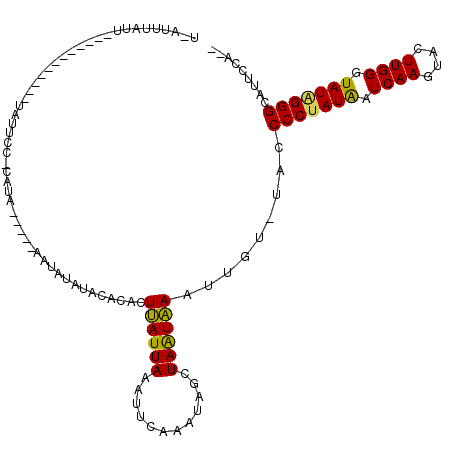
>2L_DroMel_CAF1 14237596 104 - 22407834 ----UAAUU------------UAUUCCACAUAUAAGCAAUAUAAGCACACUUAUUAAAUCCAAUUAGCUAGUAAAUUGUUUACCCCUAUAAUCAAGUACUUGGUUAUUGGGCAUUCCAGA ----.....------------...........((((((((((.(((....................))).)))..))))))).(((.((((((((....)))))))).)))......... ( -16.55) >DroSec_CAF1 70284 98 - 1 U-AUUUAUU------------UAUUCC-CAUA-----AAUAUUUACACACUUAUUAAAUUCAAUUACCUAAUAAAUUGU-UACCCCUAUAAUCAAGUACUUGGGUAUAGGGCAUUCCA-- (-(..((((------------(((...-.)))-----))))..))...............(((((........))))).-...(((((((..(((....)))..))))))).......-- ( -14.20) >DroSim_CAF1 62534 98 - 1 U-AUUUAUU------------UAUUCC-CAUA-----AAUAUUUACACACUUAUUAAACUCAAAUAGCUAAUAAAUUGU-UACCCCUAUAAUCAAGUACUUGGGUAUAGGGCAUUCCA-- (-(..((((------------(((...-.)))-----))))..))...................((((.........))-)).(((((((..(((....)))..))))))).......-- ( -13.90) >DroEre_CAF1 71293 107 - 1 CGGUUUCUUAAGAUGGAAAGUAAGACA-UAGA-----AAUAAAGACACACUAAUUAGCUAAUAAUAGCUAAUUAAUAGU-UACCCCUAUGAUCAAGUACUUGGCUAUGGGGCAU------ .(.(((((......))))).)..(((.-((..-----..)).........((((((((((....))))))))))...))-)..(((((((..(((....)))..)))))))...------ ( -24.70) >consensus U_AUUUAUU____________UAUUCC_CAUA_____AAUAUAUACACACUUAUUAAAUUCAAAUAGCUAAUAAAUUGU_UACCCCUAUAAUCAAGUACUUGGGUAUAGGGCAUUCCA__ ..................................................((((((............)))))).........(((((((.((((....)))).)))))))......... (-11.00 = -10.38 + -0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:01 2006