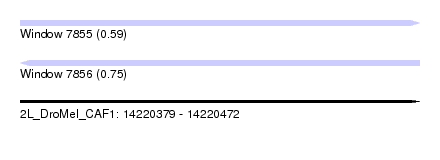
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,220,379 – 14,220,472 |
| Length | 93 |
| Max. P | 0.751487 |

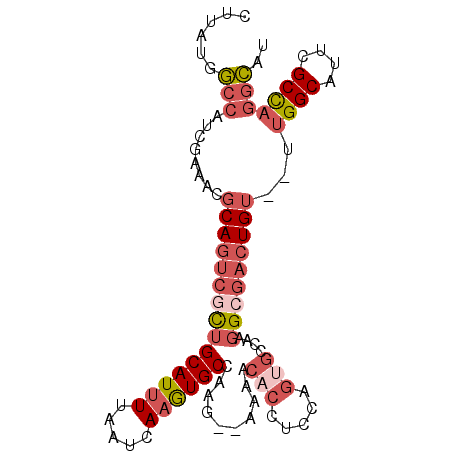
| Location | 14,220,379 – 14,220,472 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.96 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
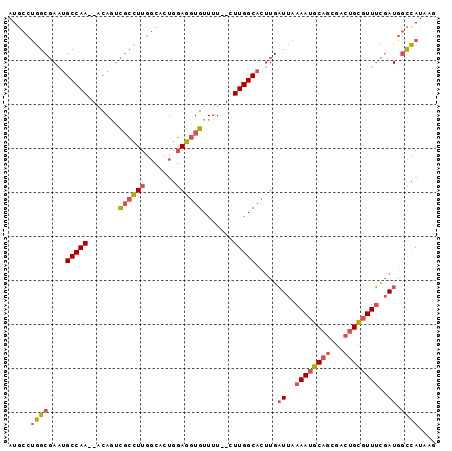
>2L_DroMel_CAF1 14220379 93 + 22407834 AUGCCUGGCGAAUGCCAA--ACAGUCGCCUUGGCACUGGAGGUGUUUU--CUUGGCACUUGAUUAAAAUGCAGCGACUGCGUUUCGAUGGCCAUAAG .((((.(((((.((....--.)).)))))..)))).((((((((((..--...))))))).((..((((((((...))))))))..))..))).... ( -32.10) >DroGri_CAF1 42740 90 + 1 AUACCAAGCCAGCGCCAAGUGCAAGUGCUUUUC------AAUUGUUGUUGCUUGGCAUGUGCUUAAAAUGCUAAGUAUGUUUUG-UAUUUUUAUUGU ((((.((((....(((((((((((.((.....)------).))))....)))))))..(((((((......))))))))))).)-)))......... ( -20.50) >DroSec_CAF1 53234 93 + 1 AUGCCUGGCGAAUGCCAA--ACAGUCGCCUUGGCACUGGAGGUGUUUU--CUUGGCACUUGAUUAAAAUGCAGCGACUGCGUUUCGAUGGCCAUAAG .((((.(((((.((....--.)).)))))..)))).((((((((((..--...))))))).((..((((((((...))))))))..))..))).... ( -32.10) >DroSim_CAF1 45446 93 + 1 AUGCCUGGCGAAUGCCAA--ACAGUCUCCUUGGCACUGGAGGUGUUUU--CUUGGCACUUGAUUAAAAUGCAGCGACUGCGUUUCGAUGGCCAUAAG ..(((...(((.((((((--(((.(((((........))))))))...--.)))))).)))((..((((((((...))))))))..)))))...... ( -29.90) >DroEre_CAF1 54310 93 + 1 AUGCCUGGCGAAUGCCAA--ACAGUCGCCUUGGCUCUGGAGGUGUUUU--CUUGGCACUUGAUUAAAAUGCAGCGACUGCGUUUCGAUGGCCAUAAG ..(((.(((((.((....--.)).)))))..)))..((((((((((..--...))))))).((..((((((((...))))))))..))..))).... ( -30.90) >DroYak_CAF1 42490 93 + 1 AUGCCUGGCGAAUGCCAA--ACAGUCGCCUUGGCACUGGAGGUGUUUU--CUUGGCACUUGAUUAAAAUGCAGCGACUGCGUUUCGAUGGCCAUAAG .((((.(((((.((....--.)).)))))..)))).((((((((((..--...))))))).((..((((((((...))))))))..))..))).... ( -32.10) >consensus AUGCCUGGCGAAUGCCAA__ACAGUCGCCUUGGCACUGGAGGUGUUUU__CUUGGCACUUGAUUAAAAUGCAGCGACUGCGUUUCGAUGGCCAUAAG .....((((....(((((.......((((((.......)))))).......))))).....((..((((((((...))))))))..)).)))).... (-21.26 = -21.96 + 0.70)

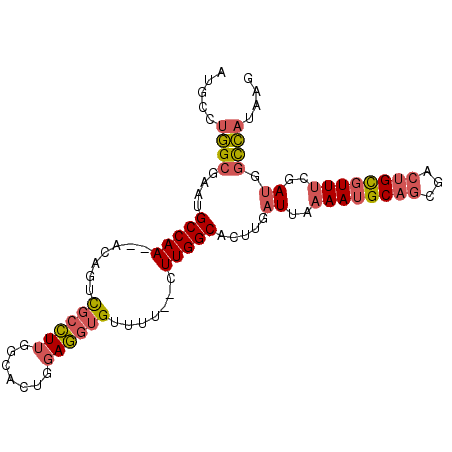
| Location | 14,220,379 – 14,220,472 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -19.74 |
| Energy contribution | -21.55 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14220379 93 - 22407834 CUUAUGGCCAUCGAAACGCAGUCGCUGCAUUUUAAUCAAGUGCCAAG--AAAACACCUCCAGUGCCAAGGCGACUGU--UUGGCAUUCGCCAGGCAU ......(((........(((((((((((((((.....))))))....--....(((.....)))....)))))))))--.((((....))))))).. ( -30.20) >DroGri_CAF1 42740 90 - 1 ACAAUAAAAAUA-CAAAACAUACUUAGCAUUUUAAGCACAUGCCAAGCAACAACAAUU------GAAAAGCACUUGCACUUGGCGCUGGCUUGGUAU ............-......(((((.(((......(((....(((((((((......))------)....((....)).))))))))).))).))))) ( -17.60) >DroSec_CAF1 53234 93 - 1 CUUAUGGCCAUCGAAACGCAGUCGCUGCAUUUUAAUCAAGUGCCAAG--AAAACACCUCCAGUGCCAAGGCGACUGU--UUGGCAUUCGCCAGGCAU ......(((........(((((((((((((((.....))))))....--....(((.....)))....)))))))))--.((((....))))))).. ( -30.20) >DroSim_CAF1 45446 93 - 1 CUUAUGGCCAUCGAAACGCAGUCGCUGCAUUUUAAUCAAGUGCCAAG--AAAACACCUCCAGUGCCAAGGAGACUGU--UUGGCAUUCGCCAGGCAU ......(((........((((((...((((((.....))))))....--........(((........)))))))))--.((((....))))))).. ( -25.10) >DroEre_CAF1 54310 93 - 1 CUUAUGGCCAUCGAAACGCAGUCGCUGCAUUUUAAUCAAGUGCCAAG--AAAACACCUCCAGAGCCAAGGCGACUGU--UUGGCAUUCGCCAGGCAU ......(((........(((((((((((((((.....))))))....--.......((....))....)))))))))--.((((....))))))).. ( -28.00) >DroYak_CAF1 42490 93 - 1 CUUAUGGCCAUCGAAACGCAGUCGCUGCAUUUUAAUCAAGUGCCAAG--AAAACACCUCCAGUGCCAAGGCGACUGU--UUGGCAUUCGCCAGGCAU ......(((........(((((((((((((((.....))))))....--....(((.....)))....)))))))))--.((((....))))))).. ( -30.20) >consensus CUUAUGGCCAUCGAAACGCAGUCGCUGCAUUUUAAUCAAGUGCCAAG__AAAACACCUCCAGUGCCAAGGCGACUGU__UUGGCAUUCGCCAGGCAU ......(((........(((((((((((((((.....))))))..........(((.....)))....)))))))))...((((....))))))).. (-19.74 = -21.55 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:50 2006