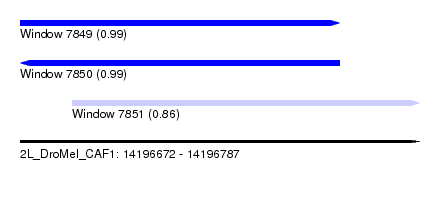
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,196,672 – 14,196,787 |
| Length | 115 |
| Max. P | 0.994545 |

| Location | 14,196,672 – 14,196,764 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -23.64 |
| Consensus MFE | -18.86 |
| Energy contribution | -20.38 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.985972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14196672 92 + 22407834 CCCUGAGCUCUGGCCCCAAAUAGACAGUCUCUCUAUCCAAAGCUUAUGGAGGGUGUUUUUAACCCACUCAGGAUUUACACUCUUCAGACUAA ...((((((.(((......(((((.......)))))))).))))))((((((((((....((.((.....)).)).))))))))))...... ( -23.50) >DroSec_CAF1 29607 92 + 1 CCCUGAGCUCUGGCCCCAAAUAGACAGCCUCUCUAUCCAAAGCCUAUGGAGGGUGUUUUUAACCCACUCAGGAUUAACACUCUCCAGAAUAA ......(((.(((......(((((.......)))))))).)))...(((((((((((.....((......))...)))))))))))...... ( -23.60) >DroSim_CAF1 21795 92 + 1 CCCUGAGCUCUGGCCCCAAAUAGACAGCCUCUCUAUCCAAAGCCUAUGGAGGGUGUUUUUAACCCACUCAGGAUUAACACUCUCCAGAAUAA ......(((.(((......(((((.......)))))))).)))...(((((((((((.....((......))...)))))))))))...... ( -23.60) >DroEre_CAF1 29305 91 + 1 CCCUGGGCUCUGGCCCCAAAUAGACAGCCUCUCUAUCCAAAGCCUAUAGAGGUUUAUUUUAACCCACUGAGGAUUUA-GCUCUCCAGACUAA ...((((((.(((......(((((.......)))))))).))))))(((.((((......))))..(((..((....-..))..))).))). ( -20.60) >DroYak_CAF1 18354 91 + 1 CCCUGGGCUCUGGCCCCAAAUAGACAUCCUCUCUAUCCAAAGCCUAUGGAGGGGGAUGUUAACCCACUGAGGACUAACACCCUAUACACUA- ((.((((((.(((......(((((.......)))))))).)))))).))..((((.(((((..((.....))..)))))))))........- ( -26.90) >consensus CCCUGAGCUCUGGCCCCAAAUAGACAGCCUCUCUAUCCAAAGCCUAUGGAGGGUGUUUUUAACCCACUCAGGAUUAACACUCUCCAGACUAA ...((((((.(((......(((((.......)))))))).))))))(((((((((((.....((......))...)))))))))))...... (-18.86 = -20.38 + 1.52)


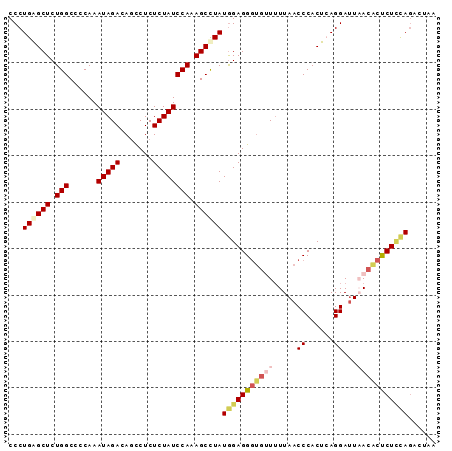
| Location | 14,196,672 – 14,196,764 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -25.90 |
| Energy contribution | -27.30 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14196672 92 - 22407834 UUAGUCUGAAGAGUGUAAAUCCUGAGUGGGUUAAAAACACCCUCCAUAAGCUUUGGAUAGAGAGACUGUCUAUUUGGGGCCAGAGCUCAGGG .((.((....)).)).....(((((((((((.......)))).......((((..((((((.......))))))..))))....))))))). ( -28.60) >DroSec_CAF1 29607 92 - 1 UUAUUCUGGAGAGUGUUAAUCCUGAGUGGGUUAAAAACACCCUCCAUAGGCUUUGGAUAGAGAGGCUGUCUAUUUGGGGCCAGAGCUCAGGG .(((((....))))).....(((((((((((.......))))......(((((..((((((.......))))))..)))))...))))))). ( -34.40) >DroSim_CAF1 21795 92 - 1 UUAUUCUGGAGAGUGUUAAUCCUGAGUGGGUUAAAAACACCCUCCAUAGGCUUUGGAUAGAGAGGCUGUCUAUUUGGGGCCAGAGCUCAGGG .(((((....))))).....(((((((((((.......))))......(((((..((((((.......))))))..)))))...))))))). ( -34.40) >DroEre_CAF1 29305 91 - 1 UUAGUCUGGAGAGC-UAAAUCCUCAGUGGGUUAAAAUAAACCUCUAUAGGCUUUGGAUAGAGAGGCUGUCUAUUUGGGGCCAGAGCCCAGGG ....(((((.(..(-(.(((((.....)))))................(((((..((((((.......))))))..)))))))..)))))). ( -28.90) >DroYak_CAF1 18354 91 - 1 -UAGUGUAUAGGGUGUUAGUCCUCAGUGGGUUAACAUCCCCCUCCAUAGGCUUUGGAUAGAGAGGAUGUCUAUUUGGGGCCAGAGCCCAGGG -.........((((((((..((.....))..))))))))((((.....(((((..((((((.......))))))..))))).......)))) ( -34.80) >consensus UUAGUCUGGAGAGUGUUAAUCCUGAGUGGGUUAAAAACACCCUCCAUAGGCUUUGGAUAGAGAGGCUGUCUAUUUGGGGCCAGAGCUCAGGG ....................(((((((((((.......))))......(((((..((((((.......))))))..)))))...))))))). (-25.90 = -27.30 + 1.40)

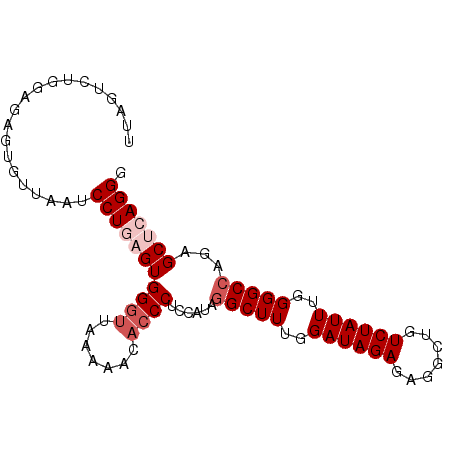
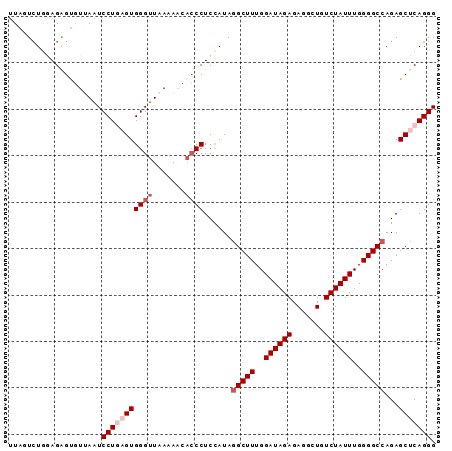
| Location | 14,196,687 – 14,196,787 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -10.66 |
| Energy contribution | -11.94 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14196687 100 + 22407834 CCAAAUAGACAGUCUCUCUAUCCAAAGCUUAUGGAGGGUGUUUUUAACCCACUCAGGAUUUACACUCUUCAGACUAACAAACAAAGAUCUAGUAAUAU-----------------GU ..........((((...((......))....((((((((((....((.((.....)).)).)))))))))))))).......................-----------------.. ( -15.10) >DroSec_CAF1 29622 86 + 1 CCAAAUAGACAGCCUCUCUAUCCAAAGCCUAUGGAGGGUGUUUUUAACCCACUCAGGAUUAACACUCUCCAGAAUAACAAA--------------UAU-----------------AU ....(((((.......)))))..........(((((((((((.....((......))...)))))))))))..........--------------...-----------------.. ( -18.20) >DroSim_CAF1 21810 86 + 1 CCAAAUAGACAGCCUCUCUAUCCAAAGCCUAUGGAGGGUGUUUUUAACCCACUCAGGAUUAACACUCUCCAGAAUAACAAA--------------UAU-----------------AU ....(((((.......)))))..........(((((((((((.....((......))...)))))))))))..........--------------...-----------------.. ( -18.20) >DroEre_CAF1 29320 99 + 1 CCAAAUAGACAGCCUCUCUAUCCAAAGCCUAUAGAGGUUUAUUUUAACCCACUGAGGAUUUA-GCUCUCCAGACUAACAAGCAAAGAUCUAUGAUUAU-----------------GU ....(((((((((((((.((.........)).)))))))............(((..((....-..))..))).............).)))))......-----------------.. ( -14.20) >DroYak_CAF1 18369 115 + 1 CCAAAUAGACAUCCUCUCUAUCCAAAGCCUAUGGAGGGGGAUGUUAACCCACUGAGGACUAACACCCUAUACACUA--AAACAAAGAUAUAUAUAUAUGUAUAUACCAUAUAUACGU .......((((((((((...((((.......)))))))))))))).........(((........)))........--.............((((((((.......))))))))... ( -23.20) >consensus CCAAAUAGACAGCCUCUCUAUCCAAAGCCUAUGGAGGGUGUUUUUAACCCACUCAGGAUUAACACUCUCCAGACUAACAAACAAAGAU_UA____UAU_________________GU ....(((((.......)))))..........(((((((((((.....((......))...))))))))))).............................................. (-10.66 = -11.94 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:46 2006