| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,482,690 – 1,482,799 |
| Length | 109 |
| Max. P | 0.821545 |

| Location | 1,482,690 – 1,482,799 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
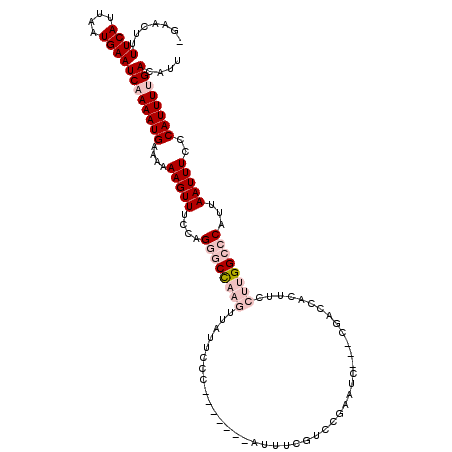
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -16.18 |
| Energy contribution | -18.24 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
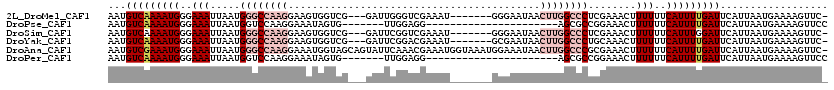
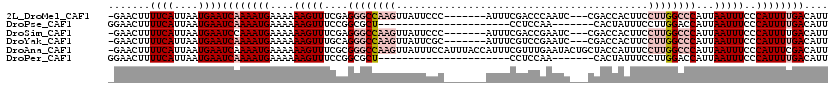
>2L_DroMel_CAF1 1482690 109 + 22407834 AAUGUCAAAAUGGGAAAUUAAUGGGCCAAGGAAGUGGUCG---GAUUGGGUCGAAAU-------GGGAAUAACUUGGCCCUCGAAACUUUUUUCAUUUUGAUUCAUUAAUGAAAAGUUC- ...(((((((((..(((.....((((((((.......(((---(......))))...-------........))))))))........)))..))))))))).................- ( -29.29) >DroPse_CAF1 135056 91 + 1 AAUGUCAAAAUGGGAAAUUAAUGGUCCAAGGAAAUAGUG-------UUGGAGG----------------------AGCGCCGGAAACUUUUUUCAUUUUGAUUCAUUAAUGAAAAGUUCC ...(((((((((..(((.......(((((.(......).-------)))))((----------------------....))(....).)))..))))))))).................. ( -18.90) >DroSim_CAF1 102820 109 + 1 AAUGUCAAAAUGGGAAAUUAAUGGGCCAAGGAAGUGGUCG---GAUUCGGUCGAAAU-------GGGAAUAACUUGGCCCUCGAAACUUUUUUCAUUUGGAUUCAUUAAUGAAAAGUUC- ...(((.(((((..(((.....((((((((....((..((---....))..))....-------........))))))))........)))..))))).))).................- ( -26.61) >DroYak_CAF1 103541 109 + 1 AAUGUCAAAAUGGGAAAUUAAUGGGCCAAGGAAGUGGUCG---GAUUCGGACGAAAU-------GCGAAUAACUUGGCCCUGCAAACUUUUUUCAUUUUGAUUCAUUAAUGAAAAGUUC- ...(((((((((..(((.....((((((((....((.(((---.((((....).)))-------.))).)).))))))))........)))..))))))))).................- ( -32.02) >DroAna_CAF1 101518 119 + 1 AAUGUCGAAAUGGGAAAUUAAUGGGCCAAGGAAAUGGUAGCAGUAUUCAAACGAAAUGGUAAAUGGAAAUAACUUGGCCCGCGAAACUUUUUUCAUUUUGAUUCAUUAAUGAAAAGUUC- ...(((((((((..(((((..(((((((((.......((.((...(((....))).)).)).((....))..)))))))))..))...)))..))))))))).................- ( -28.80) >DroPer_CAF1 133178 91 + 1 AAUGUCAAAAUGGGAAAUUAAUGGUCCAAGGAAAUAGUG-------UUGGAGG----------------------AGCGCCGGAAACUUUUUUCAUUUUGAUUCAUUAAUGAAAAGUUCC ...(((((((((..(((.......(((((.(......).-------)))))((----------------------....))(....).)))..))))))))).................. ( -18.90) >consensus AAUGUCAAAAUGGGAAAUUAAUGGGCCAAGGAAAUGGUCG___GAUUCGGACGAAAU_______GGGAAUAACUUGGCCCUCGAAACUUUUUUCAUUUUGAUUCAUUAAUGAAAAGUUC_ ...(((((((((..(((.....((((((((..........................................))))))))........)))..))))))))).................. (-16.18 = -18.24 + 2.06)
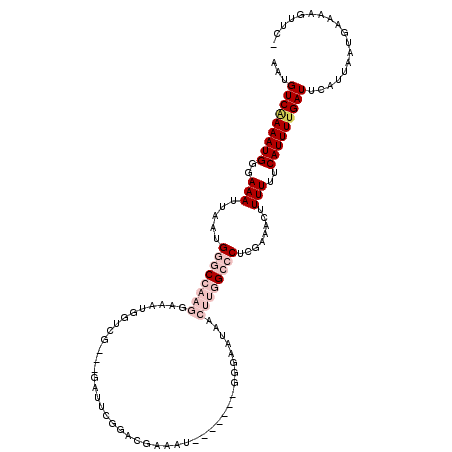
| Location | 1,482,690 – 1,482,799 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -17.68 |
| Consensus MFE | -11.67 |
| Energy contribution | -13.45 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1482690 109 - 22407834 -GAACUUUUCAUUAAUGAAUCAAAAUGAAAAAAGUUUCGAGGGCCAAGUUAUUCCC-------AUUUCGACCCAAUC---CGACCACUUCCUUGGCCCAUUAAUUUCCCAUUUUGACAUU -......((((....))))((((((((....(((((....((((((((........-------...(((........---))).......))))))))...)))))..)))))))).... ( -21.77) >DroPse_CAF1 135056 91 - 1 GGAACUUUUCAUUAAUGAAUCAAAAUGAAAAAAGUUUCCGGCGCU----------------------CCUCCAA-------CACUAUUUCCUUGGACCAUUAAUUUCCCAUUUUGACAUU (((((((((((((..........)))))..)))))))).((....----------------------..(((((-------..........)))))))...................... ( -10.10) >DroSim_CAF1 102820 109 - 1 -GAACUUUUCAUUAAUGAAUCCAAAUGAAAAAAGUUUCGAGGGCCAAGUUAUUCCC-------AUUUCGACCGAAUC---CGACCACUUCCUUGGCCCAUUAAUUUCCCAUUUUGACAUU -((((((((((((..((....)))))))..)))))))...((((((((........-------..(((....)))..---..........))))))))...................... ( -20.45) >DroYak_CAF1 103541 109 - 1 -GAACUUUUCAUUAAUGAAUCAAAAUGAAAAAAGUUUGCAGGGCCAAGUUAUUCGC-------AUUUCGUCCGAAUC---CGACCACUUCCUUGGCCCAUUAAUUUCCCAUUUUGACAUU -......((((....))))((((((((....(((((....((((((((..(.(((.-------..(((....)))..---)))....)..))))))))...)))))..)))))))).... ( -24.40) >DroAna_CAF1 101518 119 - 1 -GAACUUUUCAUUAAUGAAUCAAAAUGAAAAAAGUUUCGCGGGCCAAGUUAUUUCCAUUUACCAUUUCGUUUGAAUACUGCUACCAUUUCCUUGGCCCAUUAAUUUCCCAUUUCGACAUU -((((((((((((..........)))))..)))))))...((((((((..(..............(((....)))............)..))))))))...................... ( -19.27) >DroPer_CAF1 133178 91 - 1 GGAACUUUUCAUUAAUGAAUCAAAAUGAAAAAAGUUUCCGGCGCU----------------------CCUCCAA-------CACUAUUUCCUUGGACCAUUAAUUUCCCAUUUUGACAUU (((((((((((((..........)))))..)))))))).((....----------------------..(((((-------..........)))))))...................... ( -10.10) >consensus _GAACUUUUCAUUAAUGAAUCAAAAUGAAAAAAGUUUCCAGGGCCAAGUUAUUCCC_______AUUUCGUCCGAAUC___CGACCACUUCCUUGGCCCAUUAAUUUCCCAUUUUGACAUU .......((((....))))((((((((....(((((....((((((((..........................................))))))))...)))))..)))))))).... (-11.67 = -13.45 + 1.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:29 2006