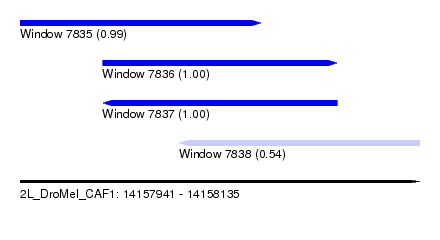
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,157,941 – 14,158,135 |
| Length | 194 |
| Max. P | 0.999921 |

| Location | 14,157,941 – 14,158,058 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -62.43 |
| Consensus MFE | -42.11 |
| Energy contribution | -43.23 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14157941 117 + 22407834 CCCAUCACGCACGCAGUUCCCGCUCCUGGAAGUGCGGUGGCCGCUGCUGCAGCCGCGGCUGCGGCCGCAGCGGUGGUCGCUGCCACAAGUUCCAGUGCCGGCGGCACCAGCACCACG ............((.....(((((((((((((((((((((((((((((((.(((((....))))).)))))))))))))))).)))...)))))).)..))))).....))...... ( -62.00) >DroWil_CAF1 343715 117 + 1 CCCAUAACCCACGCAGUUCCAGCUCCUGGCAGUGCAGUGGCCGCGGCAGCAGCUGCUGCGGCUGCUGCAGCAGUUGCAGCUGCCACCAGCUCCGGAGCUGGAGGCACUAGCACUACA ............(((((((((((((((((((((((((..((((((((((...))))))))))..)))).((....)).)))))))........)))))))))...))).))...... ( -64.20) >DroYak_CAF1 260968 117 + 1 CCCAUCACGCACGCGGUUCCCGCUCCUGGAAGUGCGGUGGCUGCUGCAGCAGCCGCGGCUGCGGCCGCAGCUGUAGUGGCUGCCACAAGUUCCACUGCCGGCGGGACCAGCACCACU ............(((((.((((((..(((((.(...(((((.((..(.(((((.(((((....))))).))))).)..)).))))).).))))).....))))))))).))...... ( -61.10) >consensus CCCAUCACGCACGCAGUUCCCGCUCCUGGAAGUGCGGUGGCCGCUGCAGCAGCCGCGGCUGCGGCCGCAGCAGUAGUAGCUGCCACAAGUUCCAGUGCCGGCGGCACCAGCACCACA ...........((((.((((.......)))).))))(((((.((.(((((.((.(((((....))))).)).))))).)).)))))........((((.((.....)).)))).... (-42.11 = -43.23 + 1.12)

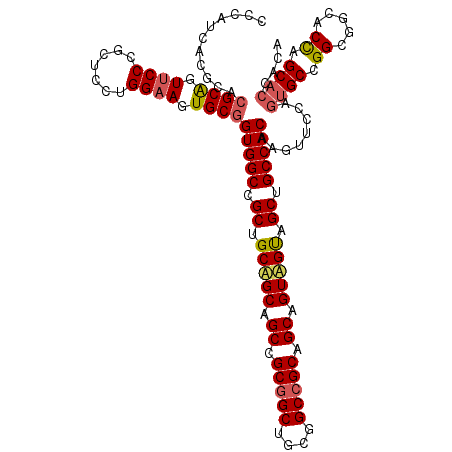
| Location | 14,157,981 – 14,158,095 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -60.93 |
| Consensus MFE | -42.49 |
| Energy contribution | -40.74 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14157981 114 + 22407834 CCGCUGCUGCAGCCGCGGCUGCGGCCGCAGCGGUGGUCGCUGCCACAAGUUCCAGUGCCGGCGGCACCAGCACCACGGCUGCCGGCAAUGCCACAGCUACCUGCGGCAUGGGCG ((((((((((.(((((....))))).)))))))))).....(((...........((((((((((............))))))))))(((((.(((....))).))))).))). ( -65.50) >DroWil_CAF1 343755 107 + 1 CCGCGGCAGCAGCUGCUGCGGCUGCUGCAGCAGUUGCAGCUGCCACCAGCUCCGGAGCUGGAGGCACUAGCACUACAACGGCUGGCAAUGCCACAGUGACUUGUGGU------- (((((((.(((((((((((((...))))))))))))).)))))..((((((....)))))).))..(((((.(......))))))....(((((((....)))))))------- ( -55.30) >DroYak_CAF1 261008 114 + 1 CUGCUGCAGCAGCCGCGGCUGCGGCCGCAGCUGUAGUGGCUGCCACAAGUUCCACUGCCGGCGGGACCAGCACCACUGCCGCCGGCAAUGCCACUGCUACCUGCGGCAUGGGCG ..(((...(((((....))))).(((((((..((((((((.......((.....))((((((((...(((.....)))))))))))...))))))))...)))))))...))). ( -62.00) >consensus CCGCUGCAGCAGCCGCGGCUGCGGCCGCAGCAGUAGUAGCUGCCACAAGUUCCAGUGCCGGCGGCACCAGCACCACAGCCGCCGGCAAUGCCACAGCUACCUGCGGCAUGGGCG ..((.(((((....))))).)).(((((((..(((((.(((......)))..((.(((((((((..............))))))))).)).....))))))))))))....... (-42.49 = -40.74 + -1.75)

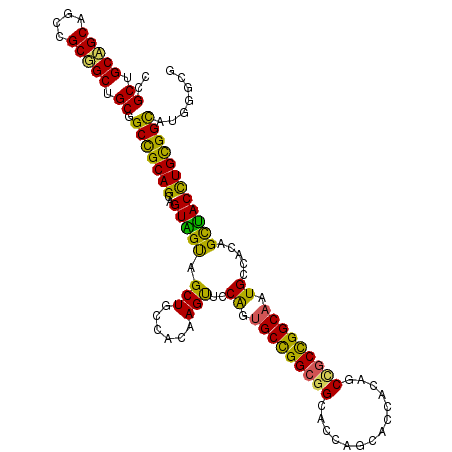
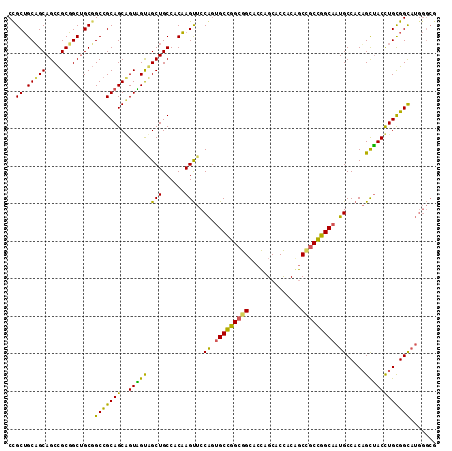
| Location | 14,157,981 – 14,158,095 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -62.67 |
| Consensus MFE | -45.32 |
| Energy contribution | -47.33 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.72 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14157981 114 - 22407834 CGCCCAUGCCGCAGGUAGCUGUGGCAUUGCCGGCAGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCGACCACCGCUGCGGCCGCAGCCGCGGCUGCAGCAGCGG (((...((((((((((.((((((((...))).)))))...((((((((.....))))))))...))))))))))))).((.(.(((((((((((....))))))))))).).)) ( -68.80) >DroWil_CAF1 343755 107 - 1 -------ACCACAAGUCACUGUGGCAUUGCCAGCCGUUGUAGUGCUAGUGCCUCCAGCUCCGGAGCUGGUGGCAGCUGCAACUGCUGCAGCAGCCGCAGCAGCUGCUGCCGCGG -------...........((((((((.(((.(((.((((((((.....((((.((((((....)))))).)))))))))))).))))))(((((.......))))))))))))) ( -55.20) >DroYak_CAF1 261008 114 - 1 CGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCAGUGGUGCUGGUCCCGCCGGCAGUGGAACUUGUGGCAGCCACUACAGCUGCGGCCGCAGCCGCGGCUGCUGCAGCAG ......((((((((((.((....))((((((((((((((.....)))...)))))))))))...))))))))))((..(..((((((((((....))))))))))..)..)).. ( -64.00) >consensus CGCCCAUGCCGCAGGUAGCUGUGGCAUUGCCGGCAGCAGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCCACAACAGCUGCGGCCGCAGCCGCGGCUGCUGCAGCGG .....(((((((((....)))))))))(((((((((((((((((((((.....)))))..........((.(((((.......))))).))....)))))))))))))..))). (-45.32 = -47.33 + 2.01)

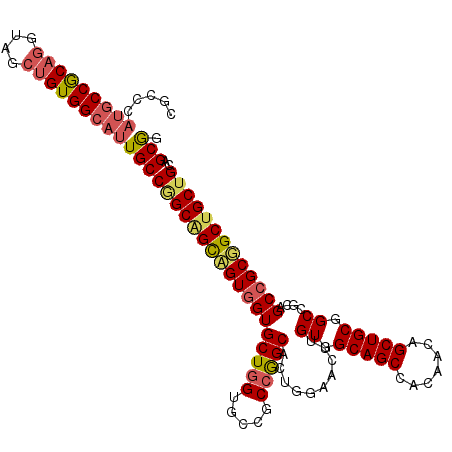
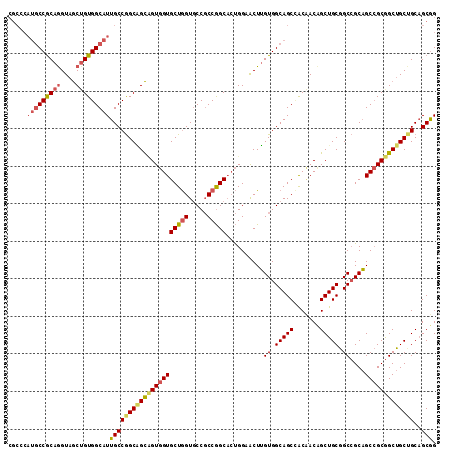
| Location | 14,158,018 – 14,158,135 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -61.50 |
| Consensus MFE | -43.46 |
| Energy contribution | -44.10 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14158018 117 - 22407834 GCGGCUGCCGCGCUGGUCAGUCCGGUGGCAUCCCCCACACCGCCCAUGCCGCAGGUAGCUGUGGCAUUGCCGGCAGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCG ...(((((((((....((((((((((((((((.(((((.(.(((.(((((((((....)))))))))....))).)..)))).)..))))))))))).)))))....))))))))). ( -69.10) >DroSec_CAF1 246778 117 - 1 GCGGCUGCUGCGCUGGUCAGACCGGUAGCAUCCCCAACACCGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCC ..((((((..((....((((.((((..(((...(((.((((((....(((((.((((((....))..)))).))))).)))))).))))))..))))..))))....))..)))))) ( -61.40) >DroSim_CAF1 243152 117 - 1 GCGGCUGCUGCGCUGGUCAGACCGGUAGCAUCCCCAACACCGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCC ..((((((..((....((((.((((..(((...(((.((((((....(((((.((((((....))..)))).))))).)))))).))))))..))))..))))....))..)))))) ( -61.40) >DroYak_CAF1 261045 117 - 1 GCGGCUGCCGCGCUGGUCAGUCCGGUGGCAUCCCCCACGCCGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCAGUGGUGCUGGUCCCGCCGGCAGUGGAACUUGUGGCAGCC ..((((((((((....(((.((((((((.(((.(((((((((((.(((((((........)))))))....)))))).)))).)..))).))))))).).)))....)))))))))) ( -66.20) >DroAna_CAF1 241747 117 - 1 GCAGCCGCAACGCUGGUGAGUCCGGCGGCAUCCGCAGCUCCACCCACGCCGCAGGUGACGGUUGCGUUGCCAGCGGUCGUUGUGCUGGUUCCUCCGGCUCCUGAGCUGGUGGCAGCC ((.(((((..((((((.....)))))).......((((((.....(((((((.((..(((....)))..)).)))).)))...(((((.....)))))....))))))))))).)). ( -57.90) >DroPer_CAF1 313577 117 - 1 GCGGCCGCCACAUUUGUGAGGCCGGUGGCAUCCGCGACUCCUGCUCCUCCGCAGGUGACGGUGGCAUUGCCCGCGGACGUGGUGCUCGUUCCGCCGGCACCAGAGCUGGUAGCAGCC .(((((..(((....))).)))))((.((....)).))..(((((((..(...((((.((((((.((.((((((....)))).))..)).)))))).))))...)..)).))))).. ( -53.00) >consensus GCGGCUGCCGCGCUGGUCAGUCCGGUGGCAUCCCCAACACCGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCC ..((((((((((((((.....)))))....(((..............(((((.(((((........))))).)))))...((((((((.....)))))))))))....))))))))) (-43.46 = -44.10 + 0.64)

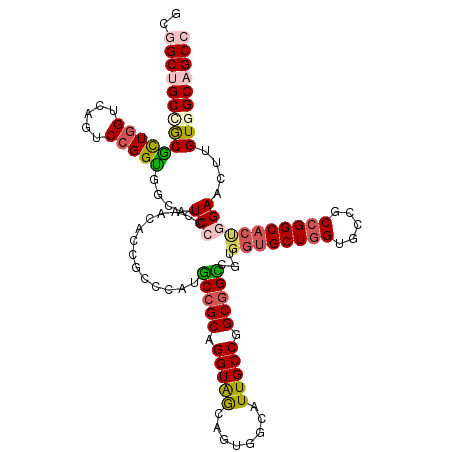
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:33 2006