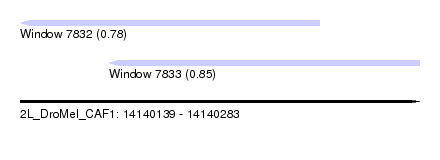
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,140,139 – 14,140,283 |
| Length | 144 |
| Max. P | 0.846996 |

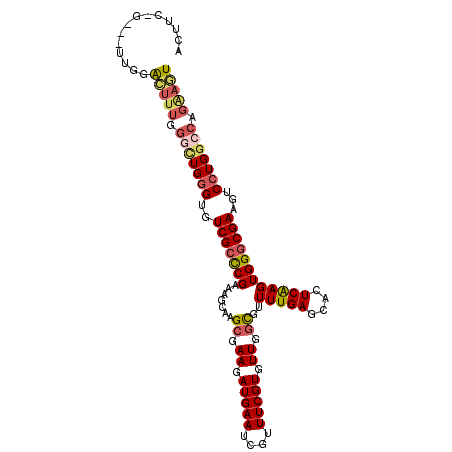
| Location | 14,140,139 – 14,140,247 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.77 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -31.51 |
| Energy contribution | -32.27 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14140139 108 - 22407834 ACUUCAGUCUUUGGACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGAGCACUCAAGUGGGCGAAGUCCUGGCCAGAAGU (((((((((....))))...(((..((..(((((((.......((.((.(((((....))))).)).))..(((((....))))))))))))...))..))).))))) ( -45.10) >DroSec_CAF1 228954 101 - 1 ACUUCAG-------ACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGAGCACUCAAGUGGGCGAAGUCCUGGCCAGAAGU .......-------(((((.(((..((..(((((((.......((.((.(((((....))))).)).))..(((((....))))))))))))...))..))).))))) ( -41.50) >DroSim_CAF1 225415 101 - 1 ACUUCAG-------ACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGAGCACUCAAGUGGGCGAAGUCCUGGCCAGAAGU .......-------(((((.(((..((..(((((((.......((.((.(((((....))))).)).))..(((((....))))))))))))...))..))).))))) ( -41.50) >DroYak_CAF1 242921 107 - 1 ACUCC-GACUUUGGACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGAGCACUCAAGUGGGCGAAGUCCUGGCCAGAAGU ..(((-(....))))((((.(((..((..(((((((.......((.((.(((((....))))).)).))..(((((....))))))))))))...))..))).)))). ( -42.40) >DroAna_CAF1 224850 98 - 1 ----------UUGGGGUGGGGAAUGGGUCUCGGCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGUGUUUUGAGCACUCAAGUGGGCGAAGUCCUGGCCAGGACU ----------....(((.((((.....)))).)))....((..((.((.(((((....))))).))(((((.....)))))...))..))..((((((....)))))) ( -29.00) >DroPer_CAF1 291719 105 - 1 G--UG-GGUUCUGGGUCUUGGAUUGGGUGUCGCUCGAAAGCACGAGAAGAUGAAUCGUUUCGUUUUGGCGUUUUGAGCACUCGAGUGGGCGAAGUCGUGGGGAGGACU .--..-((((((...(((((((((..((.((((((((..(((((..((((((((....))))))))..))).....))..))))))))))..)))).))))))))))) ( -30.50) >consensus ACUUC_G___UUGGACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGAGCACUCAAGUGGGCGAAGUCCUGGCCAGAAGU ..............(((((.(((((((..(((((((.......((.((.(((((....))))).)).))..(((((....))))))))))))...))))))).))))) (-31.51 = -32.27 + 0.75)


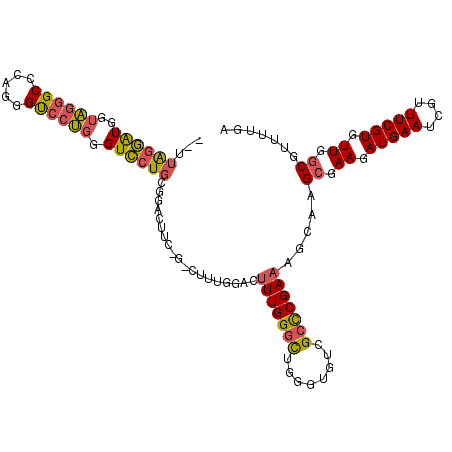
| Location | 14,140,171 – 14,140,283 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14140171 112 - 22407834 --UUAGGAUGGUAGGGCCCAGGGUCCUGGGUCCUGCGGACUUCAGUCUUUGGACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGA --...((...(((((((((((....))))))))))).(((((((((((.........)))))))).)))..)).(((((..((.((.(((((....))))).)).))))))).. ( -46.00) >DroSec_CAF1 228986 105 - 1 --UUAGGAUGGUAGGGCCCAGGGUCCUGGGUCCUGCGGACUUCAG-------ACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGA --(((((((.(((((((((((....)))))))))))........(-------..(((((((........)))))))..)..((.((.(((((....))))).)).))))))))) ( -43.70) >DroSim_CAF1 225447 105 - 1 --UUAGGAUGGUAGGGCCCAGGGUCCUGGGUCCUGCGGACUUCAG-------ACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGA --(((((((.(((((((((((....)))))))))))........(-------..(((((((........)))))))..)..((.((.(((((....))))).)).))))))))) ( -43.70) >DroEre_CAF1 231879 104 - 1 --UUGGGAUGGUUGGG-------CCCAGAGUCCUGCGGACUCC-GACUUUGGACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGA --(((((.(((((.((-------((((((((((.((((...))-).)...)))))))))))).))..))))))))......((.((.(((((....))))).)).))....... ( -41.90) >DroYak_CAF1 242953 111 - 1 --AUAGGAUGGUUGGGCUGAGGGCCGAGGGUCCUGCGGACUCC-GACUUUGGACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGA --.((((((((((((((((((((((...)))))).))).).))-))))...(..(((((((........)))))))..)..((.((.(((((....))))).)).)))))))). ( -38.90) >DroPer_CAF1 291751 96 - 1 CUGUGCUGUGGC------------U---GGUGCUUUGGG--UG-GGUUCUGGGUCUUGGAUUGGGUGUCGCUCGAAAGCACGAGAAGAUGAAUCGUUUCGUUUUGGCGUUUUGA ....(((....(------------(---.((((((((((--((-(...((.((((...)))).))..)))))).))))))).))((((((((....)))))))))))....... ( -30.10) >consensus __UUAGGAUGGUAGGGCCCAGGGUCCUGGGUCCUGCGGACUUC_G_CUUUGGACUUUGGGCUGGGUGUCGCCCGAAAGCAAGCGAAGAUGAAUCGUUUCGUGUUGGCGUUUUGA ...((((((..((((((.....)))))).))))))...................(((((((........))))))).....((.((.(((((....))))).)).))....... (-24.12 = -24.25 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:29 2006