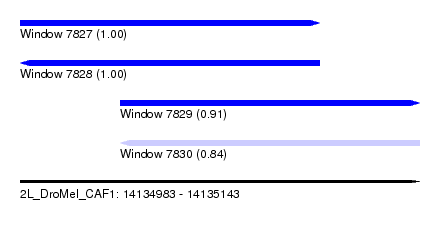
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,134,983 – 14,135,143 |
| Length | 160 |
| Max. P | 0.999970 |

| Location | 14,134,983 – 14,135,103 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -44.77 |
| Consensus MFE | -42.63 |
| Energy contribution | -43.13 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
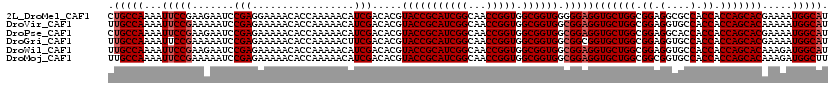
>2L_DroMel_CAF1 14134983 120 + 22407834 CUGCCAAAAUUCCGAAGAAUCCGAGGAAAACACCAAAAACAUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGGGGAGGUGCUGGCGGAGGCGCCACCACCAGCACGAAAAUGGCAU .(((((...((((...........((......)).............(.(((((((((((...))))).)))))).)))))(((((((.((.(....).)).))))))).....))))). ( -45.80) >DroVir_CAF1 386935 120 + 1 UUGCCAAAAUUCCGAAAAAUCCGAGAAAAACACCAAAAACAUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGAGGUGCCACCACCAGCACAAAAAUGGCAU .(((((...(((((.......(((.................))).....(((((((((((...))))).)))))).)))))(((((((.((.(....).)).))))))).....))))). ( -44.93) >DroPse_CAF1 278833 120 + 1 CUGCCAAAAUUCCGAAGAAUCCGAGAAAAACACCAAAAACAUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGAGGCACCACCACCAGCACGAAAAUGGCAU .(((((...(((((.......(((.................))).....(((((((((((...))))).)))))).)))))(((((((.((.(....).)).))))))).....))))). ( -44.83) >DroGri_CAF1 265553 120 + 1 UUGCCAAAAUUCCGAAAAAUCCGAGAAAAACACCAAAAACUUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGCGGUGCUGGCGGAGGUGCCACCACCAGCACGAAAAUGGCAU .(((((...(((((.......)).)))...............((...(((((((((((((...))))).))))).))).))(((((((.((.(....).)).))))))).....))))). ( -43.60) >DroWil_CAF1 317119 120 + 1 UUGCCAAAAUUCCGAAGAAUCCGAGAAAAACACCAAAAACAUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGAGGUGCCACCACCAGCACAAAGAUGGCAU .(((((...(((((.......(((.................))).....(((((((((((...))))).)))))).)))))(((((((.((.(....).)).))))))).....))))). ( -44.93) >DroMoj_CAF1 345904 120 + 1 UUGCCAAAAUUCCGAAAAAUCCGAGAAAAACACCAAAAACAUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGCGGUGCCACCACCAGCACAAAGAUGGCUU ..((((...(((((.......(((.................))).....(((((((((((...))))).)))))).)))))(((((((.((.(....).)).))))))).....)))).. ( -44.53) >consensus UUGCCAAAAUUCCGAAAAAUCCGAGAAAAACACCAAAAACAUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGAGGUGCCACCACCAGCACAAAAAUGGCAU .(((((...(((((.......(((.................))).....(((((((((((...))))).)))))).)))))(((((((.((.(....).)).))))))).....))))). (-42.63 = -43.13 + 0.50)
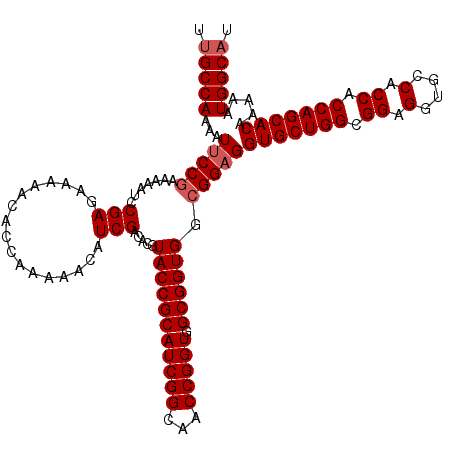
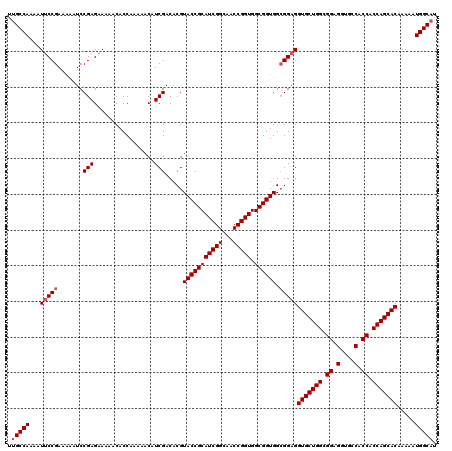
| Location | 14,134,983 – 14,135,103 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -48.83 |
| Consensus MFE | -46.19 |
| Energy contribution | -46.44 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.74 |
| SVM RNA-class probability | 0.999944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14134983 120 - 22407834 AUGCCAUUUUCGUGCUGGUGGUGGCGCCUCCGCCAGCACCUCCCCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAUGUUUUUGGUGUUUUCCUCGGAUUCUUCGGAAUUUUGGCAG .(((((.....((((((((((.(....).))))))))))............((((((..(((((((((...)))))))..))..))))))........((((((....))))))))))). ( -47.60) >DroVir_CAF1 386935 120 - 1 AUGCCAUUUUUGUGCUGGUGGUGGCACCUCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAUGUUUUUGGUGUUUUUCUCGGAUUUUUCGGAAUUUUGGCAA .(((((.((((((((((((((.(....).)))))))))).((((.......((((((..(((((((((...)))))))..))..)))))).......))))......))))...))))). ( -47.14) >DroPse_CAF1 278833 120 - 1 AUGCCAUUUUCGUGCUGGUGGUGGUGCCUCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAUGUUUUUGGUGUUUUUCUCGGAUUCUUCGGAAUUUUGGCAG .(((((.((((((((((((((.(....).)))))))))).((((.......((((((..(((((((((...)))))))..))..)))))).......))))......))))...))))). ( -49.94) >DroGri_CAF1 265553 120 - 1 AUGCCAUUUUCGUGCUGGUGGUGGCACCUCCGCCAGCACCGCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAAGUUUUUGGUGUUUUUCUCGGAUUUUUCGGAAUUUUGGCAA .(((((.((((((((((((((.(....).))))))))))((((((.((((....)))).))(((((((...)))))))........)))).................))))...))))). ( -49.10) >DroWil_CAF1 317119 120 - 1 AUGCCAUCUUUGUGCUGGUGGUGGCACCUCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAUGUUUUUGGUGUUUUUCUCGGAUUCUUCGGAAUUUUGGCAA .(((((.....((((((((((.(....).)))))))))).((((.......((((((..(((((((((...)))))))..))..)))))).......))))(((....)))...))))). ( -49.44) >DroMoj_CAF1 345904 120 - 1 AAGCCAUCUUUGUGCUGGUGGUGGCACCGCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAUGUUUUUGGUGUUUUUCUCGGAUUUUUCGGAAUUUUGGCAA ..((((.....((((((((((((....)))))))))))).((((.......((((((..(((((((((...)))))))..))..)))))).......)))).............)))).. ( -49.74) >consensus AUGCCAUUUUCGUGCUGGUGGUGGCACCUCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAUGUUUUUGGUGUUUUUCUCGGAUUCUUCGGAAUUUUGGCAA .(((((..(((((((((((((.(....).)))))))))).((((.......((((((..(((((((((...)))))))..))..)))))).......)))).......)))...))))). (-46.19 = -46.44 + 0.25)


| Location | 14,135,023 – 14,135,143 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -46.97 |
| Consensus MFE | -41.93 |
| Energy contribution | -42.43 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14135023 120 + 22407834 AUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGGGGAGGUGCUGGCGGAGGCGCCACCACCAGCACGAAAAUGGCAUCUAAAAGAAAAGCCUCGGUGUUGCUGCACAAGGUGAGUUU ...(((((.(((((((((((...))))).))))))((((..(((((((.((.(....).)).)))))))..........((.....))....)))).)))))(((.(((...)))))).. ( -42.90) >DroVir_CAF1 386975 120 + 1 AUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGAGGUGCCACCACCAGCACAAAAAUGGCAUCUAAAAGAAAAGCAUCGGUGUUGCUGCACAAGGUGAGUUC ...(((...((((.((((((...))))))((((..(((..((((((..(..((((((((...............))))))))....)...))))))..)))..))))....)))).))). ( -44.76) >DroGri_CAF1 265593 120 + 1 UUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGCGGUGCUGGCGGAGGUGCCACCACCAGCACGAAAAUGGCAUCUAAAAGAAAAGCAUCGGUGUUGCUGCACAAGGUGAGUUC ...(((...((((.((((((...))))))((((..(((.(((((((..(..((((((((...............))))))))....)...))))))).)))..))))....)))).))). ( -48.46) >DroWil_CAF1 317159 120 + 1 AUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGAGGUGCCACCACCAGCACAAAGAUGGCAUCUAAAAGAAAAGCCUCGGUGUUGCUGCACAAGGUGAGUGU ....((((.((((.((((((...))))))((((..(((...(((((((.((.(....).)).)))))))...((.(((.((.....))...)))))..)))..))))....)))).)))) ( -49.80) >DroMoj_CAF1 345944 120 + 1 AUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGCGGUGCCACCACCAGCACAAAGAUGGCUUCUAAAAGAAAAGCCUCGGUGUUGCUGCACAAGGUGAGUUC ...(((...((((.((((((...))))))((((..(((...(((((((.((.(....).)).)))))))...((.(((((.........)))))))..)))..))))....)))).))). ( -48.10) >DroAna_CAF1 220008 120 + 1 AUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGAGGCGCCACCACCAGCACGAAAAUGGCAUCUAAAAGAAAAGCCUCGGUGUUGCUGCACAAGGUGAGUGC .....(((.((((.((((((...))))))((((..(((..((((((((.((.(....).)).)))))))......(((.((.....))...))).)..)))..))))....)))).))). ( -47.80) >consensus AUCGACACGUACCGCAUCGGCAACCGGUGGCGGUGGCGGAGGUGCUGGCGGAGGUGCCACCACCAGCACAAAAAUGGCAUCUAAAAGAAAAGCCUCGGUGUUGCUGCACAAGGUGAGUUC ......((.((((.((((((...))))))((((..(((..((((((((.((.(....).)).)))))))......(((.((.....))...))).)..)))..))))....)))).)).. (-41.93 = -42.43 + 0.50)


| Location | 14,135,023 – 14,135,143 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -42.63 |
| Consensus MFE | -38.27 |
| Energy contribution | -38.93 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14135023 120 - 22407834 AAACUCACCUUGUGCAGCAACACCGAGGCUUUUCUUUUAGAUGCCAUUUUCGUGCUGGUGGUGGCGCCUCCGCCAGCACCUCCCCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAU .......(((((((......)).)))))...........(((((.......((((((((((.(....).)))))))))).......(((((...(((...)))..)))))..)))))... ( -40.20) >DroVir_CAF1 386975 120 - 1 GAACUCACCUUGUGCAGCAACACCGAUGCUUUUCUUUUAGAUGCCAUUUUUGUGCUGGUGGUGGCACCUCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAU ...........(((......)))((((((...((.....))((((......((((((((((.(....).)))))))))).((.((.((((....)))).)).))...)))).)))))).. ( -39.10) >DroGri_CAF1 265593 120 - 1 GAACUCACCUUGUGCAGCAACACCGAUGCUUUUCUUUUAGAUGCCAUUUUCGUGCUGGUGGUGGCACCUCCGCCAGCACCGCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAA ...........(((......)))((((((...((.....))((((......((((((((((.(....).)))))))))).((.((.((((....)))).))....)))))).)))))).. ( -39.30) >DroWil_CAF1 317159 120 - 1 ACACUCACCUUGUGCAGCAACACCGAGGCUUUUCUUUUAGAUGCCAUCUUUGUGCUGGUGGUGGCACCUCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAU ....((((..(((((.(((.....(((((...((.....)).))).))...((((((((((.(....).)))))))))).((.((.((((....)))).)).))))).))))).)).)). ( -42.50) >DroMoj_CAF1 345944 120 - 1 GAACUCACCUUGUGCAGCAACACCGAGGCUUUUCUUUUAGAAGCCAUCUUUGUGCUGGUGGUGGCACCGCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAU ....((((..(((((.(((.....(((((((((.....))))))).))...((((((((((((....)))))))))))).((.((.((((....)))).)).))))).))))).)).)). ( -48.90) >DroAna_CAF1 220008 120 - 1 GCACUCACCUUGUGCAGCAACACCGAGGCUUUUCUUUUAGAUGCCAUUUUCGUGCUGGUGGUGGCGCCUCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAU ((((.......)))).(((((.....(((...((.....)).))).....((.(.(((((((((((.....(....).....)))))))))))))))))))(((((((...))))))).. ( -45.80) >consensus GAACUCACCUUGUGCAGCAACACCGAGGCUUUUCUUUUAGAUGCCAUUUUCGUGCUGGUGGUGGCACCUCCGCCAGCACCUCCGCCACCGCCACCGGUUGCCGAUGCGGUACGUGUCGAU ....((((..(((((.(((.......(((...((.....)).)))......((((((((((.(....).)))))))))).((.((.((((....)))).)).))))).))))).)).)). (-38.27 = -38.93 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:26 2006