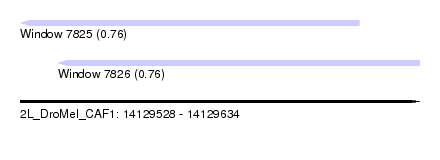
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,129,528 – 14,129,634 |
| Length | 106 |
| Max. P | 0.762312 |

| Location | 14,129,528 – 14,129,618 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.37 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
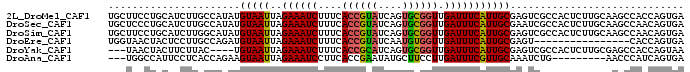
>2L_DroMel_CAF1 14129528 90 - 22407834 CCAUAUGUAAUUAGAAAUCUUUCACCGUAUCAGUGCGGUUGAUUUCAUUGCGAGUCGCCACUCUUGCAAGCCACCAGUGAGCA--UAAAAAA ...(((((.....((((((....((((((....)))))).)))))).(((((((........)))))))...........)))--))..... ( -23.90) >DroSec_CAF1 218287 90 - 1 CCAUAUGUAAUUAGAAAUCUUUCACCGUAUCAGUGCGGUUGAUUUCAUUGCGAAUCGCCACUCUUGCAAGCCAACAGUGAGCA--UAAAAAA .....((((((..((((((....((((((....)))))).))))))))))))....((((((.(((.....))).)))).)).--....... ( -22.70) >DroSim_CAF1 214763 90 - 1 GCAUAUGUAAUUAGAAAUCUUUCACCGUAUCAGUGCGGUUGAUUUCAUUGCGAGUCGCCACUCUUGCAAGCCAACAGUGAGCA--UAAAAAA ...(((((.....((((((....((((((....)))))).)))))).(((((((........)))))))...........)))--))..... ( -23.90) >DroEre_CAF1 221010 74 - 1 CCAGAUGUAAUUAGAAAUCUUUCACCGUAUCAAUGUGGUUGAUUUCAUUGCGAGU----------------CACCAGUGAGUA--UAAAAAA .....((((((..((((((..((((.........))))..))))))))))))(.(----------------((....))).).--....... ( -13.40) >DroYak_CAF1 231953 88 - 1 C----UGUAAUUAGAAAUCUUUCACCGCAUCAGUGCGGUUGAUUUCAUUGCGAGUCGCCACUCUUGCGAGCCACCAGUAAGUAUAUAAAAAA (----((......((((((....((((((....)))))).)))))).(((((((........))))))).....)))............... ( -23.20) >DroAna_CAF1 215635 81 - 1 CCAGAAGUAAUUAGAAAUCCUUCACCGAAUAUGCUUCCUUGAUUUCGUUGCAAAUCUG---------AACCCAUCAGUGAUUA--UAAAAAA ......(((((((((((((.....................)))))).........(((---------(.....))))))))))--)...... ( -8.50) >consensus CCAUAUGUAAUUAGAAAUCUUUCACCGUAUCAGUGCGGUUGAUUUCAUUGCGAGUCGCCACUCUUGCAAGCCACCAGUGAGCA__UAAAAAA ......(((((..((((((....((((((....)))))).)))))))))))......................................... (-12.61 = -12.37 + -0.25)



| Location | 14,129,538 – 14,129,634 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.37 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14129538 96 - 22407834 UGCUUCCUGCAUCUUGCCAUAUGUAAUUAGAAAUCUUUCACCGUAUCAGUGCGGUUGAUUUCAUUGCGAGUCGCCACUCUUGCAAGCCACCAGUGA ....(((((...(((((.....(((((..((((((....((((((....)))))).)))))))))))((((....))))..)))))....))).)) ( -25.20) >DroSec_CAF1 218297 96 - 1 UGCUCCCUGCAUCUUGCCAUAUGUAAUUAGAAAUCUUUCACCGUAUCAGUGCGGUUGAUUUCAUUGCGAAUCGCCACUCUUGCAAGCCAACAGUGA (((.....)))....((.((.((((((..((((((....((((((....)))))).)))))))))))).)).))((((.(((.....))).)))). ( -23.10) >DroSim_CAF1 214773 96 - 1 UGCUUCCUGCAUCUUGGCAUAUGUAAUUAGAAAUCUUUCACCGUAUCAGUGCGGUUGAUUUCAUUGCGAGUCGCCACUCUUGCAAGCCAACAGUGA (((.....)))..(((((...........((((((....((((((....)))))).)))))).(((((((........))))))))))))...... ( -29.50) >DroEre_CAF1 221020 80 - 1 UGGUAACUACUCCUUGCCAGAUGUAAUUAGAAAUCUUUCACCGUAUCAAUGUGGUUGAUUUCAUUGCGAGU----------------CACCAGUGA ((((((.......))))))((((((((..((((((..((((.........))))..)))))))))))..))----------------)........ ( -16.80) >DroYak_CAF1 231965 89 - 1 ---UAACUACUUCUUAC----UGUAAUUAGAAAUCUUUCACCGCAUCAGUGCGGUUGAUUUCAUUGCGAGUCGCCACUCUUGCGAGCCACCAGUAA ---...........(((----((......((((((....((((((....)))))).)))))).(((((((........))))))).....))))). ( -26.60) >DroAna_CAF1 215645 84 - 1 ---UGGCCAUUCCUCACCAGAAGUAAUUAGAAAUCCUUCACCGAAUAUGCUUCCUUGAUUUCGUUGCAAAUCUG---------AACCCAUCAGUGA ---..........((((..(((((((((.(((....)))....))).))))))..((((...(((.((....))---------)))..)))))))) ( -8.80) >consensus UGCUACCUACAUCUUGCCAUAUGUAAUUAGAAAUCUUUCACCGUAUCAGUGCGGUUGAUUUCAUUGCGAGUCGCCACUCUUGCAAGCCACCAGUGA ......................(((((..((((((....((((((....)))))).)))))))))))............................. (-12.61 = -12.37 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:22 2006