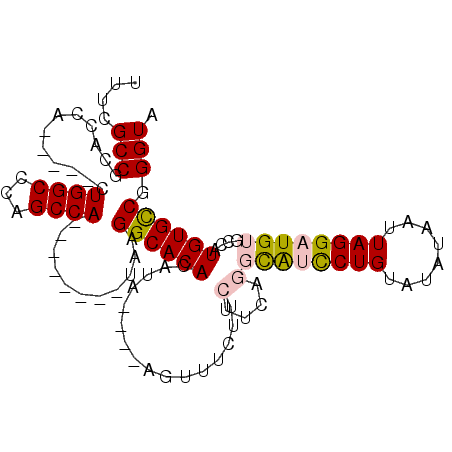
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,097,207 – 14,097,359 |
| Length | 152 |
| Max. P | 0.852742 |

| Location | 14,097,207 – 14,097,301 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
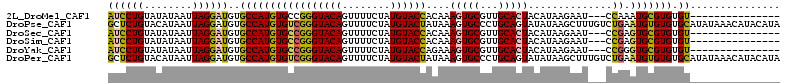
>2L_DroMel_CAF1 14097207 94 - 22407834 AUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACAAAGUGCGUUGCACUACAUAAGAAU---CCAAAUGCGUGUGU--------------- ((((((........))))))((((.(((..(..((((((........)))))).....)..))).)))).(((((...((---....))...)))))--------------- ( -26.00) >DroPse_CAF1 235815 112 - 1 GCUCUGUACAUAAUUAGGAUGUGCCAUGUGUCGGGUACAGUUUUCUAUGUACUAUAAAGUGCCCUGCAGUAUAUAAGCUUUGUCUGAAUGUGUGUGCAUAUAAACAUACAUA (((..(((((((.......((((((........))))))......)))))))((((.(.(((...))).).))))))).........((((((((........)))))))). ( -27.72) >DroSec_CAF1 186147 94 - 1 AUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACAAAGUGCGUUGCACUACAUAAGAAU---CCGAGUGCGUGUGU--------------- ((((((........))))))..((((((..(((((((((........))))))....(((((...)))))..........---.)).)..)))).))--------------- ( -28.90) >DroSim_CAF1 186911 94 - 1 AUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACAAAGUGCGUUGCACUACAUAAGAAU---CCGAGUGCGUGUGU--------------- ((((((........))))))..((((((..(((((((((........))))))....(((((...)))))..........---.)).)..)))).))--------------- ( -28.90) >DroYak_CAF1 197838 94 - 1 AUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCAGAAAGUGCGUUGCACUACAUAAGAAU---CCGGGUGCGUGUGU--------------- ((((((........))))))..((((((..((.((((((........))))))....(((((...)))))..........---...))..)))).))--------------- ( -30.40) >DroPer_CAF1 242982 112 - 1 GCUCUGUACAUAAUUAGGAUGUGCCAUGUGUCGGGUACAGUUUUCUAUGUACUAUAAAGUGCCCUGCAGUAUAUAAGCUUUGUCUGAAUGUGUGUGCAUAUAAACAUACAUA (((..(((((((.......((((((........))))))......)))))))((((.(.(((...))).).))))))).........((((((((........)))))))). ( -27.72) >consensus AUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACAAAGUGCGUUGCACUACAUAAGAAU___CCGAAUGCGUGUGU_______________ ((((((........))))))..(((((((((((((((((........))))))....(((((...)))))..............)).))))))).))............... (-21.76 = -21.10 + -0.66)
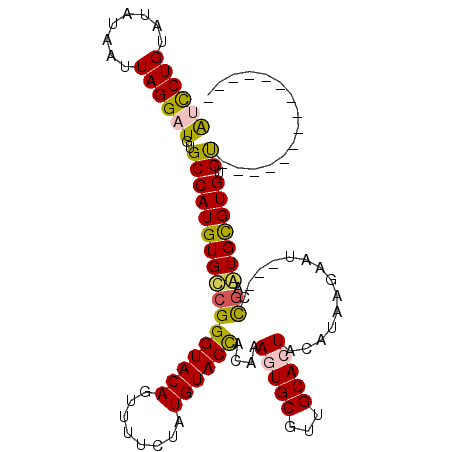
| Location | 14,097,227 – 14,097,334 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.852742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
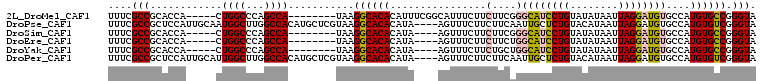
>2L_DroMel_CAF1 14097227 107 - 22407834 -------UAAGGCACACAUUUCGGCAUUUCUUCUUCGGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACAAAGUGCGUUGCACUACAU -------...((((((......(((.......(....)((((((((........))))))))))).)))))).((((((........))))))....(((((...))))).... ( -37.50) >DroPse_CAF1 235853 110 - 1 AUGCUCGUAAGGCACACAUA----AGUUUCUUCUUCAAUUGCUCUGUACAUAAUUAGGAUGUGCCAUGUGUCGGGUACAGUUUUCUAUGUACUAUAAAGUGCCCUGCAGUAUAU (((((.(((.(((((....(----((......)))..........(((((((.......((((((........))))))......)))))))......))))).)))))))).. ( -29.52) >DroSim_CAF1 186931 103 - 1 -------UAAGGCACACAUA----AGUUUCUUCUUCGGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACAAAGUGCGUUGCACUACAU -------...((((((...(----((......))).((((((((((........)))))))).)).)))))).((((((........))))))....(((((...))))).... ( -35.50) >DroEre_CAF1 189006 103 - 1 -------UAAGGCACACAUA----AGUUUCUUCUUCUGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCAGAAAGUGCGUUGCACUACAU -------...((((((((.(----((......))).))((((((((........))))))))....)))))).((((((........))))))....(((((...))))).... ( -35.50) >DroAna_CAF1 185031 99 - 1 -------GAAGGCACACACA----AGAUUCUUCUUAUGUCGGCCUGUAUAUAAUUAGGAUGGGCCGUAAGCCG-GUACAGUUUUCUAUUUACCAUAAACUGCAA---AGUACAU -------...(((..(((.(----(((....)))).)))((((((((.((....))..))))))))...)))(-(((.(((.....)))))))....(((....---))).... ( -24.20) >DroPer_CAF1 243020 110 - 1 AUGCUCGUAAGGCACACAUA----AGUUUCUUCUUCAAUUGCUCUGUACAUAAUUAGGAUGUGCCAUGUGUCGGGUACAGUUUUCUAUGUACUAUAAAGUGCCCUGCAGUAUAU (((((.(((.(((((....(----((......)))..........(((((((.......((((((........))))))......)))))))......))))).)))))))).. ( -29.52) >consensus _______UAAGGCACACAUA____AGUUUCUUCUUCAGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCAUAAAGUGCGUUGCACUACAU ..........((((((.........((((.......))))..((((........)))).))))))........((((((........))))))....(((((...))))).... (-16.90 = -17.68 + 0.78)
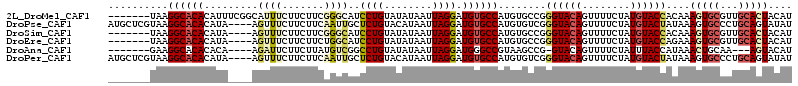
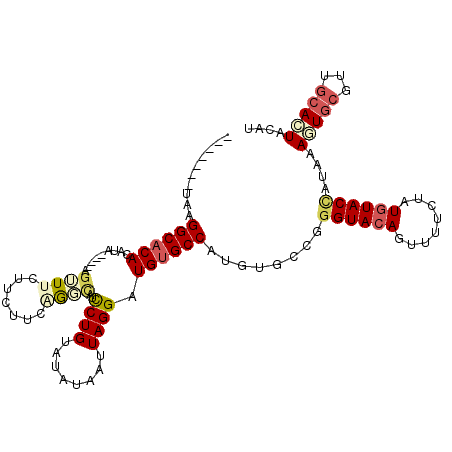
| Location | 14,097,264 – 14,097,359 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -20.27 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14097264 95 - 22407834 UUUCGCCGCACCA-----CUGGCCCAGCCA--------UAAGGCACACAUUUCGGCAUUUCUUCUUCGGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUA ....((((.....-----.))))...(((.--------...((((((......(((.......(....)((((((((........))))))))))).)))))).))). ( -31.90) >DroPse_CAF1 235890 104 - 1 UUUCGCCGCUCCAUUGCAAUGGCUUGGCCACAUGCUCGUAAGGCACACAUA----AGUUUCUUCUUCAAUUGCUCUGUACAUAAUUAGGAUGUGCCAUGUGUCGGGUA ....((((..((((....))))..))))....((((((((.((((((...(----((......))).......((((........)))).))))))...)).)))))) ( -23.70) >DroSim_CAF1 186968 91 - 1 UUUCGCCGCACCA-----CUGGCCCAGCCA--------UAAGGCACACAUA----AGUUUCUUCUUCGGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUA ....((((.....-----.))))...(((.--------...((((((...(----((......))).((((((((((........)))))))).)).)))))).))). ( -29.90) >DroEre_CAF1 189043 91 - 1 UUUCGCCGCACCA-----CUGGCCCAGCCA--------UAAGGCACACAUA----AGUUUCUUCUUCUGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUA ....((((.....-----.))))...(((.--------...((((((((.(----((......))).))((((((((........))))))))....)))))).))). ( -29.90) >DroYak_CAF1 197895 91 - 1 UUUCGCCGCACCA-----CUGGCCCAGCCA--------UAAGGCACACAUA----AGUUUCUUCUGCUGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUA ....(((((((..-----.(((((((((..--------.((((.((.....----.)).))))..)))))(((((((........))))))).)))).))))..))). ( -30.80) >DroPer_CAF1 243057 104 - 1 UUUCGCCGCUCCAUUGCAUUGGCUUGGCCACAUGCUCGUAAGGCACACAUA----AGUUUCUUCUUCAAUUGCUCUGUACAUAAUUAGGAUGUGCCAUGUGUCGGGUA ....((((..(((......)))..))))....((((((((.((((((...(----((......))).......((((........)))).))))))...)).)))))) ( -23.60) >consensus UUUCGCCGCACCA_____CUGGCCCAGCCA________UAAGGCACACAUA____AGUUUCUUCUUCAGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUA ....(((............((((...))))...........((((((................(....)((((((((........))))))))....)))))).))). (-20.27 = -20.38 + 0.11)
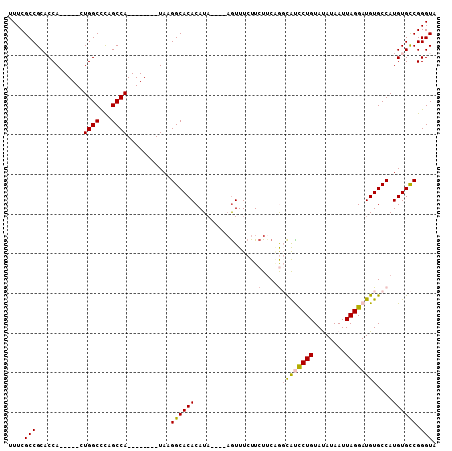
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:17 2006