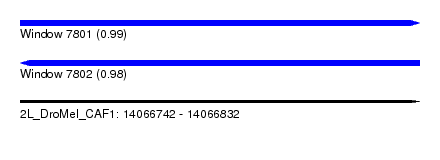
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,066,742 – 14,066,832 |
| Length | 90 |
| Max. P | 0.985049 |

| Location | 14,066,742 – 14,066,832 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 71.65 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -12.83 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
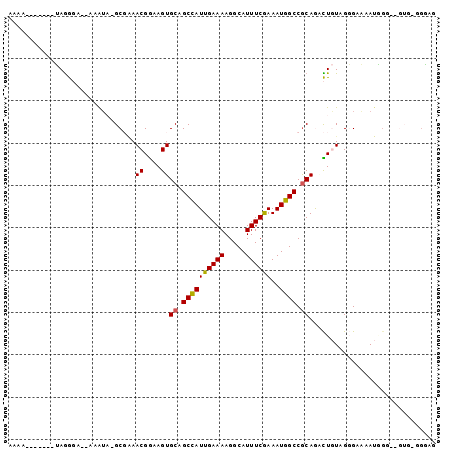
>2L_DroMel_CAF1 14066742 90 + 22407834 AAAAAA-----UAGGGA--AAAUA-GCGAAACGGAAGUGCAGCCAUUGAAAAGGCAUUUCGAAAUGGCCGCAGACUGUAGGGAAAAUGGU--GUA-UGAAG .....(-----(((...--.....-.....((....))((.((((((((((.....))))))..)))).))...))))............--...-..... ( -15.80) >DroVir_CAF1 280336 76 + 1 AC-------A--AAACCAAAAGGCAGCC-AACGGAAGUGCAGCCAUUGAAAAGGCAUUUCAAAAUGGCCGCAAUUUAUGGUGGCAA--------------- ..-------.--..........((.(((-(((....))((.((((((((((.....)))))..))))).))......)))).))..--------------- ( -22.00) >DroGri_CAF1 186558 89 + 1 GC----CGUAAGAGACC--AAGUC-GCCGAACGAAAGUGCAGCUAUUGAAAAGGCAUUUCAAAAUGGCCACAAUUUAUGGUGGCAACUGC--CUG-GGC-- ((----(.........(--((...-((.(.((....)).).))..)))...(((((...(.....)(((((........)))))...)))--)).-)))-- ( -23.10) >DroSec_CAF1 155488 92 + 1 AAAAAA-----UAGGGA--AAAUA-GCGAAACGGAAGUGCAGCCAUUGAAAAGGCAUUUCGAAAUGGCCGCAGGCUGUAGGGAAAAUGGGGAGUG-GGGAG ......-----......--..(((-((...((....))((.((((((((((.....))))))..)))).))..))))).................-..... ( -19.10) >DroSim_CAF1 156408 92 + 1 AAAAAA-----UAGGGA--AAAUA-GCGAAACGGAAGUGCAGCCAUUGAAAAGGCAUUUCGAAAUGGCCGCAGACUGUAGGGAAAAUGGGGAGUA-AGGAG .....(-----(((...--.....-.....((....))((.((((((((((.....))))))..)))).))...)))).................-..... ( -15.80) >DroEre_CAF1 158323 90 + 1 AAAA------AUAGGGA--AAAUA-GCGAAACGGAAGUGCAGCCAUUGAAAAGGCAUUUCGAAAUGGCCGCAGACUGUUGGGAAAAUAGA--GGGGGGGCG ....------.....((--((...-((...((....)))).(((........))).))))......(((.(...(((((.....))))).--...).))). ( -19.50) >consensus AAAA_______UAGGGA__AAAUA_GCGAAACGGAAGUGCAGCCAUUGAAAAGGCAUUUCGAAAUGGCCGCAGACUGUAGGGAAAAUGGG__GUG_GGGAG ..............................((....))((.((((((((((.....))))))..)))).)).............................. (-12.83 = -12.63 + -0.19)

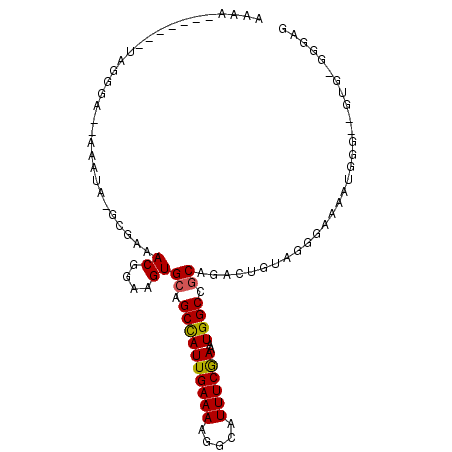
| Location | 14,066,742 – 14,066,832 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 71.65 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -14.97 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14066742 90 - 22407834 CUUCA-UAC--ACCAUUUUCCCUACAGUCUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCACUUCCGUUUCGC-UAUUU--UCCCUA-----UUUUUU .....-...--..................((((((((((..((((.....))))))))))))))............-.....--......-----...... ( -16.30) >DroVir_CAF1 280336 76 - 1 ---------------UUGCCACCAUAAAUUGCGGCCAUUUUGAAAUGCCUUUUCAAUGGCUGCACUUCCGUU-GGCUGCCUUUUGGUUU--U-------GU ---------------..((((........(((((((((..(((((.....)))))))))))))).......)-))).(((....)))..--.-------.. ( -24.06) >DroGri_CAF1 186558 89 - 1 --GCC-CAG--GCAGUUGCCACCAUAAAUUGUGGCCAUUUUGAAAUGCCUUUUCAAUAGCUGCACUUUCGUUCGGC-GACUU--GGUCUCUUACG----GC --(((-.((--(((((((((.....(((.((..((....((((((.....))))))..))..)).))).....)))-)))).--.)))).....)----)) ( -25.00) >DroSec_CAF1 155488 92 - 1 CUCCC-CACUCCCCAUUUUCCCUACAGCCUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCACUUCCGUUUCGC-UAUUU--UCCCUA-----UUUUUU .....-.......................((((((((((..((((.....))))))))))))))............-.....--......-----...... ( -16.30) >DroSim_CAF1 156408 92 - 1 CUCCU-UACUCCCCAUUUUCCCUACAGUCUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCACUUCCGUUUCGC-UAUUU--UCCCUA-----UUUUUU .....-.......................((((((((((..((((.....))))))))))))))............-.....--......-----...... ( -16.30) >DroEre_CAF1 158323 90 - 1 CGCCCCCCC--UCUAUUUUCCCAACAGUCUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCACUUCCGUUUCGC-UAUUU--UCCCUAU------UUUU .........--...........(((.(..((((((((((..((((.....))))))))))))))...).)))....-.....--.......------.... ( -16.50) >consensus CUCCC_CAC__CCCAUUUUCCCUACAGUCUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCACUUCCGUUUCGC_UAUUU__UCCCUA_______UUUU .............................((((((((((..((((.....))))))))))))))..................................... (-14.97 = -15.00 + 0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:59 2006