
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,066,600 – 14,066,736 |
| Length | 136 |
| Max. P | 0.978982 |

| Location | 14,066,600 – 14,066,712 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.29 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14066600 112 + 22407834 UUCAUCUCACCCUGUCUGCCUGGCCAACGACUUGGUU--------UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUCCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCAACGG ...........((((..((..((((........(((.--------.(((...((((((.((......)).)))))))))..)))(((.((((((....))))))))))))).))..)))) ( -26.20) >DroPse_CAF1 192910 102 + 1 ----GCUGUCUCUGU----CUGGCCCAGGACUUGACU--------UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUCCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGAC-- ----((.(((...((----((......))))..))).--------.((((.((((((((.((((((.........(((......)))....)))))).)))))))).)))).))....-- ( -25.12) >DroGri_CAF1 186428 90 + 1 ----------UUUGU----G-------CGACUUCG-G--------UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUUCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGCCAA ----------.(((.----(-------((((....-)--------)((((.((((((((.((((((.........(((......)))....)))))).)))))))).))))...)))))) ( -24.12) >DroWil_CAF1 218862 112 + 1 UUUAGCU---GCUG-----UUGGCCAUUGCCUUUGCUGCCUUCGCUCGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUUCAGUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGCCAA ....((.---((..-----..((((.......((((((..(((((.......))))).))))))............((.....((((((...)))))).......)))))).)).))... ( -28.70) >DroAna_CAF1 152946 100 + 1 UUCGGCACUCUGUGU----CUGGCCAACGACUUGGCA--------UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUCCACUGCAAUAAAAUUGCAAUUUUAUGCGGCC--------G ...(((((...))))----)..(((((....))))).--------.((((.((((((((.((((((.........(((......)))....)))))).)))))))).))))--------. ( -27.92) >DroPer_CAF1 204970 102 + 1 ----GCUGUCUCUGU----CUGGCCCAGGACUUGACU--------UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUCCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGAC-- ----((.(((...((----((......))))..))).--------.((((.((((((((.((((((.........(((......)))....)))))).)))))))).)))).))....-- ( -25.12) >consensus ____GCU_UCUCUGU____CUGGCCAACGACUUGGCU________UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUCCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGAC__ ..............................................((((.((((((((.((((((.........(((......)))....)))))).)))))))).))))......... (-16.12 = -16.29 + 0.17)



| Location | 14,066,600 – 14,066,712 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -17.79 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14066600 112 - 22407834 CCGUUGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGGAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA--------AACCAAGUCGUUGGCCAGGCAGACAGGGUGAGAUGAA (((((.((.((((((.....((((((((...))))))))((....(((((.(((....))).))))).....)).--------.........).)))))..)).))).)).......... ( -27.40) >DroPse_CAF1 192910 102 - 1 --GUCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGGAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA--------AGUCAAGUCCUGGGCCAG----ACAGAGACAGC---- --((((((.....((.....((((((((...))))))))...)).(((((.(((....))).)))))....))).--------.(((..((.....))..)----))...)))...---- ( -25.80) >DroGri_CAF1 186428 90 - 1 UUGGCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGAAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA--------C-CGAAGUCG-------C----ACAAA---------- ((((((((.((..((.......((((((...))))))((((((((............))))...))))...))..--------)-)...))))-------)----.))).---------- ( -24.00) >DroWil_CAF1 218862 112 - 1 UUGGCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCACUGAAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCGAGCGAAGGCAGCAAAGGCAAUGGCCAA-----CAGC---AGCUAAA (((((.((.(((((((......)))......(((((.((...((.(((((.(((....))).)))))........((....)).))..)))))))))))..-----..))---.))))). ( -34.20) >DroAna_CAF1 152946 100 - 1 C--------GGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGGAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA--------UGCCAAGUCGUUGGCCAG----ACACAGAGUGCCGAA (--------((((((((...((((((((...))))))))((....(((((.(((....))).))))).....)))--------)))...(((........)----)).....).)))).. ( -26.20) >DroPer_CAF1 204970 102 - 1 --GUCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGGAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA--------AGUCAAGUCCUGGGCCAG----ACAGAGACAGC---- --((((((.....((.....((((((((...))))))))...)).(((((.(((....))).)))))....))).--------.(((..((.....))..)----))...)))...---- ( -25.80) >consensus __GUCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGGAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA________AGCCAAGUCGUUGGCCAG____ACAGAGA_AGC____ .........((((((.(...((((((((...)))))))).).)).(((((.(((....))).)))))............................))))..................... (-17.79 = -18.40 + 0.61)



| Location | 14,066,637 – 14,066,736 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.37 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.978982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14066637 99 + 22407834 -----UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUCCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCAACGGACAAUAACAA------CAACAAC---GG--UGGCA -----.((((.((((((((.((((((.........(((......)))....)))))).)))))))).)))).((((.((...........------......)---).--)))). ( -24.45) >DroVir_CAF1 280227 94 + 1 -----UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUUCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGCCAAACAAGCGAAG------C-----CUCCAA---A--A -----.((((((((((((.((......)).))))))((((...((((((...)))))).......((((....)))).....))))))))------)-----).....---.--. ( -26.40) >DroGri_CAF1 186443 100 + 1 -----UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUUCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGCCAAACAAGCGAAG------CAAAAACACCAA--CA--A -----(((....((((((.((......)).))))))(((((.(((((((...)))))........((((....))))......)).))))------).......))).--..--. ( -25.00) >DroWil_CAF1 218894 112 + 1 UUCGCUCGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUUCAGUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGCCAAACAACCAAAAAGUAAGAAACAAC---GAAACAACA ((((((..(((((((((((.((((((.........(((......)))....)))))).)))))))((((....))))...........))))..))......)---)))...... ( -21.42) >DroMoj_CAF1 245599 94 + 1 -----UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUUCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCAGCCGCCAAACAAGCGAAG------C-----CUCCCA---A--A -----.((((((((((((.((......)).))))))((((...((((((...)))))).......((((....)))).....))))))))------)-----).....---.--. ( -26.40) >DroAna_CAF1 152979 89 + 1 -----UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUCCACUGCAAUAAAAUUGCAAUUUUAUGCGGCC--------GACA--AACAA------CAAAAAC---AC--UACCA -----.((((.((((((((.((((((.........(((......)))....)))))).)))))))).))))--------....--.....------.......---..--..... ( -17.82) >consensus _____UGGCUUUGUGAAAUAGCAAUUACUUUUUCAUGCUUCACUGCAAUAAAAUUGCAAUUUUAUGCGGCCUGCCGCCAAACAAGCAAAA______CAA_AAC___AA___A__A ......((((.((((((((.((((((.........(((......)))....)))))).)))))))).))))............................................ (-16.20 = -16.37 + 0.17)

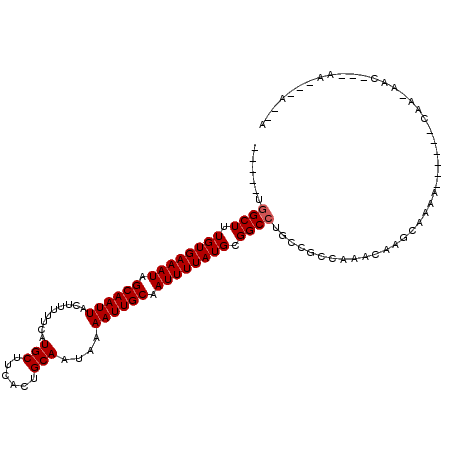
| Location | 14,066,637 – 14,066,736 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -15.74 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14066637 99 - 22407834 UGCCA--CC---GUUGUUG------UUGUUAUUGUCCGUUGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGGAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA----- (((((--.(---(......------...........)).)))))(((.((.....((((((((...))))))))...)).(((((.(((....))).)))))....))).----- ( -23.73) >DroVir_CAF1 280227 94 - 1 U--U---UUGGAG-----G------CUUCGCUUGUUUGGCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGAAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA----- .--.---.....(-----(------(((.((((.....((((....)))).....((((((((...)))))))).)))).(((((.(((....))).)))))..))))).----- ( -29.20) >DroGri_CAF1 186443 100 - 1 U--UG--UUGGUGUUUUUG------CUUCGCUUGUUUGGCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGAAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA----- .--..--.((((.....((------((((((..((((.((((....))))...))))((((((...))))))))))))))(((((.(((....))).)))))....))))----- ( -31.40) >DroWil_CAF1 218894 112 - 1 UGUUGUUUC---GUUGUUUCUUACUUUUUGGUUGUUUGGCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCACUGAAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCGAGCGAA ......(((---((......(((((((((.((.((((.((((....)))).......((((((...))))))...)))))).)))))))))(((((.........)))))))))) ( -27.70) >DroMoj_CAF1 245599 94 - 1 U--U---UGGGAG-----G------CUUCGCUUGUUUGGCGGCUGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGAAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA----- .--.---.....(-----(------(((.((((.....((((....)))).....((((((((...)))))))).)))).(((((.(((....))).)))))..))))).----- ( -29.20) >DroAna_CAF1 152979 89 - 1 UGGUA--GU---GUUUUUG------UUGUU--UGUC--------GGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGGAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA----- .(((.--((---(((((((------(.((.--....--------.)).)))....((((((((...)))))))))))))))((((.(((....))).)))).....))).----- ( -18.60) >consensus U__U___UU___GUU_UUG______CUUCGCUUGUUUGGCGGCAGGCCGCAUAAAAUUGCAAUUUUAUUGCAGUGAAGCAUGAAAAAGUAAUUGCUAUUUCACAAAGCCA_____ ............................................(((.((.(...((((((((...)))))))).).)).(((((.(((....))).)))))....)))...... (-15.74 = -15.85 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:57 2006