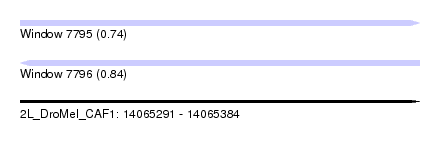
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,065,291 – 14,065,384 |
| Length | 93 |
| Max. P | 0.842558 |

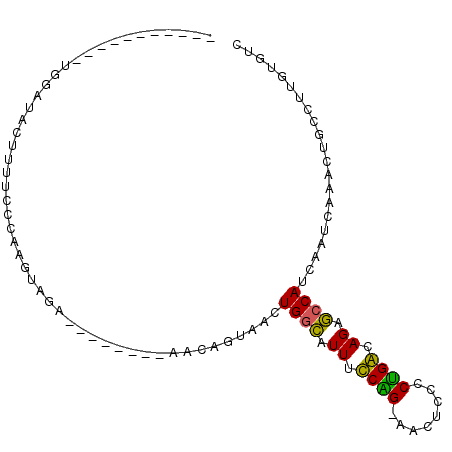
| Location | 14,065,291 – 14,065,384 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 66.53 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -9.56 |
| Energy contribution | -8.48 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14065291 93 + 22407834 GACACAAGGCAGUUUGAUUGAUGGCGCUGUCAGGGGAGUU-CUGAAAAUGCCAGUUACUGUU--------UCUACUUGAAAAAAGUAGCAGAAUACUUUGAA ....(((((..((((......(((((...(((((.....)-))))...)))))((((((.((--------((.....))))..)))))).)))).))))).. ( -22.90) >DroPse_CAF1 191326 88 + 1 GACACAAGGAAUUUUGAUUGAUGGCUCUGCCGG---AGUUGCCGGCAAUGCCAGGGCUUGUUGGAAAGCUUCUCUAUGUGAGAAGUAUCCA----------- .......(((.(((..((.(.((((..((((((---.....))))))..))))...)..))..))).(((((((.....))))))).))).----------- ( -34.80) >DroSec_CAF1 154160 93 + 1 GACACAAGGCAGUUUGAUUGAUGGCGCUGUCAGGGGAGUU-CUGAAAAUGCCAGUUACUGGU--------UCUACUGGGAAAAAGUAGCAGAAUACUUUGAA ....(((((..((((......(((((...(((((.....)-))))...)))))((((((..(--------(((....))))..)))))).)))).))))).. ( -25.40) >DroEre_CAF1 156009 93 + 1 GACAUCAGGCAGUUUGAUUGAUGGUCCUGUCAGGGGAGUU-CUGAAAAUACCAGUUACUGUU--------UCUACUUUGGAAAAGUAACCAAAUACUUUGAA ....(((((..(((((.....((((....(((((.....)-))))....))))((((((.((--------((......)))).)))))))))))..))))). ( -21.60) >DroAna_CAF1 151572 78 + 1 GACACAAGGCACUUUGAUUGAUGGCCCUGCCAGAGAAUUC-CCGGGAAUGUCAGUUACUUUG--------AAUACUUUGAAAAAAAA--------------- ....(((((.....(((((((((((...))).....((((-....)))))))))))))))))--------.................--------------- ( -13.70) >DroPer_CAF1 203380 88 + 1 GACACAAGGAAUUUUGAUUGAUGGCUCUGCCGG---AGUUGCCGGCAAUGCCAGGGCCUGUUGGAAAGCUUCUCUAUGUGAGAAGUAUCCA----------- .......(((.(((..((.(.((((..((((((---.....))))))..))))...)..))..))).(((((((.....))))))).))).----------- ( -34.70) >consensus GACACAAGGCAGUUUGAUUGAUGGCCCUGCCAGGGGAGUU_CCGAAAAUGCCAGUUACUGUU________UCUACUUGGAAAAAGUAGCCA___________ .....................((((....((((........))))....))))................................................. ( -9.56 = -8.48 + -1.08)

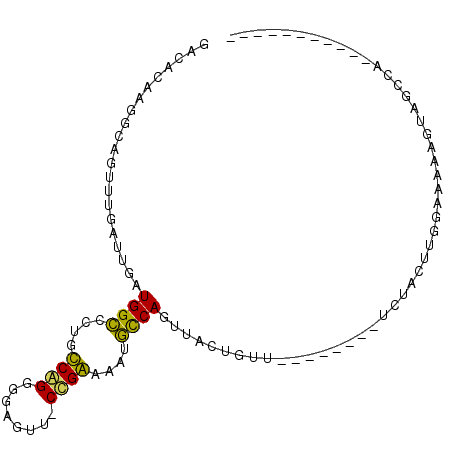
| Location | 14,065,291 – 14,065,384 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 66.53 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -8.83 |
| Energy contribution | -7.75 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14065291 93 - 22407834 UUCAAAGUAUUCUGCUACUUUUUUCAAGUAGA--------AACAGUAACUGGCAUUUUCAG-AACUCCCCUGACAGCGCCAUCAAUCAAACUGCCUUGUGUC ..(((.((((((((((..........))))))--------)........((((.((.((((-.......)))).)).))))..........))).))).... ( -16.30) >DroPse_CAF1 191326 88 - 1 -----------UGGAUACUUCUCACAUAGAGAAGCUUUCCAACAAGCCCUGGCAUUGCCGGCAACU---CCGGCAGAGCCAUCAAUCAAAAUUCCUUGUGUC -----------((((..((((((.....))))))...))))(((((...((((.(((((((.....---))))))).)))).............)))))... ( -30.39) >DroSec_CAF1 154160 93 - 1 UUCAAAGUAUUCUGCUACUUUUUCCCAGUAGA--------ACCAGUAACUGGCAUUUUCAG-AACUCCCCUGACAGCGCCAUCAAUCAAACUGCCUUGUGUC ..(((.((((((((((..........))))))--------)........((((.((.((((-.......)))).)).))))..........))).))).... ( -15.80) >DroEre_CAF1 156009 93 - 1 UUCAAAGUAUUUGGUUACUUUUCCAAAGUAGA--------AACAGUAACUGGUAUUUUCAG-AACUCCCCUGACAGGACCAUCAAUCAAACUGCCUGAUGUC .(((..(((((((((((((((((.......))--------)).))))))((((....((((-.......))))....)))).....)))).))).))).... ( -16.90) >DroAna_CAF1 151572 78 - 1 ---------------UUUUUUUUCAAAGUAUU--------CAAAGUAACUGACAUUCCCGG-GAAUUCUCUGGCAGGGCCAUCAAUCAAAGUGCCUUGUGUC ---------------.................--------..........((((...((((-(.....)))))(((((((..........).)))))))))) ( -13.10) >DroPer_CAF1 203380 88 - 1 -----------UGGAUACUUCUCACAUAGAGAAGCUUUCCAACAGGCCCUGGCAUUGCCGGCAACU---CCGGCAGAGCCAUCAAUCAAAAUUCCUUGUGUC -----------((((..((((((.....))))))...))))(((((...((((.(((((((.....---))))))).)))).............)))))... ( -30.09) >consensus ___________UGGAUACUUUUCCCAAGUAGA________AACAGUAACUGGCAUUUCCAG_AACUCCCCUGACAGAGCCAUCAAUCAAACUGCCUUGUGUC .................................................((((.((.((((........)))).)).))))..................... ( -8.83 = -7.75 + -1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:54 2006