
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,062,355 – 14,062,468 |
| Length | 113 |
| Max. P | 0.699268 |

| Location | 14,062,355 – 14,062,468 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -45.27 |
| Consensus MFE | -16.69 |
| Energy contribution | -18.03 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
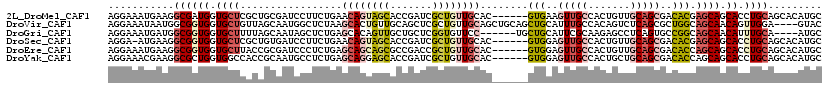
>2L_DroMel_CAF1 14062355 113 - 22407834 AGGAAAUGAAGGCGAUGGUGCUCGCUGCGAUCCUUCUGAACAGUAGCACCGAUCGCUGUUGCAC------GUGAAGUUGCCACUGUUGCAGCGACACGAGCAGCACCUGCAGCACAUGC (((.......((((((((((((.((((.((....))....)))))))))).))))))(((((.(------(((..(((((.......)))))..)))).))))).)))(((.....))) ( -45.70) >DroVir_CAF1 274476 115 - 1 AGGAAAUAAUGGCGGUGGUGCUGUUAGCAAUGGCUCUAAGCACUGUUGCAGCUCGCUGUUGCAGCUGCAGCUGCAUUUGCCACAGUCUCAGCGCUGGCAGCAACAGUUGGA----GUAC ...........((((..(((((...(((....)))...)))))..)))).((((((((((((.(((((.((((...(((...)))...)))))).))).))))))))..))----)).. ( -45.00) >DroGri_CAF1 181292 109 - 1 AGGAAAUGAUGGCGGUGGUGCUUUUAGCAAUAGCUCUGAGCACAGUUGCUGCUCGGUGUUCC------UGCUGCAUUCGCAAGAGCCUCAGUGCCGGCAGCAACAUUUGCA----AUGC ...........((((..((((((..(((....)))..)))))).((((((((.(((((...(------((..((..((....))))..))))))))))))))))..)))).----.... ( -42.20) >DroSec_CAF1 151241 112 - 1 AGGA-AUGAAGGCGGUGGUGCUCGCUGUGAUCCUUCUGAACAGUAGCACCGAUCGCUGUUGCAC------GUGGAGUUGCCACUGUUGCAGCGACACGAGCAGCACCUGCAGCACAUGC (((.-.....((((((((((((.(((((...........))))))))))).))))))(((((.(------(((..(((((.......)))))..)))).))))).)))(((.....))) ( -49.00) >DroEre_CAF1 153107 113 - 1 AGGAAAUGAAGGCGGUGGUGCUUACCGCGAUCCCUCUGAGCAGCAGCGCCGACCGCUGUUGCAC------GUGGAGUUGCCACUGUUGCAGCGACACCAGCAGCACCUGCAGCACAUGC ......((...((((((.((((..(((((.((.....))(((((((((.....))))))))).)------)))).(((((..(....)..)))))...)))).)))).))..))..... ( -43.40) >DroYak_CAF1 161576 113 - 1 AGGAAACGAAGGCGCUGGUGGCCACCGCAAUGCCUCUGAGCAGGAGCACCGAUCGCUGUUGCAC------GUGGAGUUGCCACUGCUGCAGCGACACCAGCAGCACCUGCAGCACAUGC .(....)....(((((((((.....((...(((..((....))..))).)).(((((((.(((.------((((.....))))))).)))))))))))))).((....)).))...... ( -46.30) >consensus AGGAAAUGAAGGCGGUGGUGCUCACAGCAAUCCCUCUGAGCAGUAGCACCGAUCGCUGUUGCAC______GUGGAGUUGCCACUGUUGCAGCGACACCAGCAGCACCUGCAGCACAUGC ...........((((((.((((.................((((((((.......))))))))........(((..(((((.......)))))..))).)))).)).))))......... (-16.69 = -18.03 + 1.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:51 2006