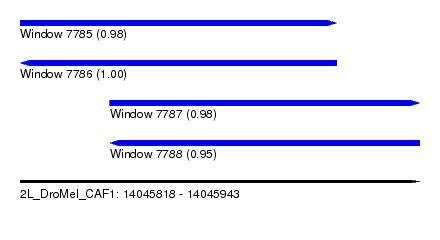
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,045,818 – 14,045,943 |
| Length | 125 |
| Max. P | 0.999955 |

| Location | 14,045,818 – 14,045,917 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14045818 99 + 22407834 CGUUUAUACAACAGUUAUACUUUGCACACCCAGUACAACUGGGAUACCCAGCACUGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCU .....((((..(((((.((((..........)))).)))))((((((...((((.((.....((........)).....)).)))).)))))))))).. ( -20.00) >DroSec_CAF1 134733 99 + 1 CUGUUAUCCAACAGUUAUACUUUGCACACCCAGUAUAACUGGGAUAACCAGCACAGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCU ..(((((((..((((((((((..........)))))))))))))))))..((((.((.....((........)).....)).))))............. ( -24.00) >DroSim_CAF1 137947 99 + 1 CUGUUAUCCAACAGUUAUACUUUGCACACCCAGUACAACUGGGAUAACCAGCACAGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCU (((((....)))))..((((..(((...(((((.....))))).......((((.((.....((........)).....)).)))).)))...)))).. ( -20.90) >DroEre_CAF1 136761 99 + 1 CUGUUAUCCAACAGUUACACUUUGCACACCCAGUACAACUGGGAUAACCGGCACAGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCU ..........((((.(((..........(((((.....)))))......(((((.((.....((........)).....)).)))))))).)))).... ( -25.70) >DroYak_CAF1 144257 99 + 1 CUGUUAUCCAACAGUUACACUUUGCACACCCAGUACAACUGGGAUAACCAGCACAGCCACAGUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCU ..........((((.(((..........(((((.....))))).......((((.((.....((........)).....)).)))).))).)))).... ( -22.50) >consensus CUGUUAUCCAACAGUUAUACUUUGCACACCCAGUACAACUGGGAUAACCAGCACAGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCU ..........((((.(((..........(((((.....))))).......((((.((.....((........)).....)).)))).))).)))).... (-20.44 = -20.20 + -0.24)

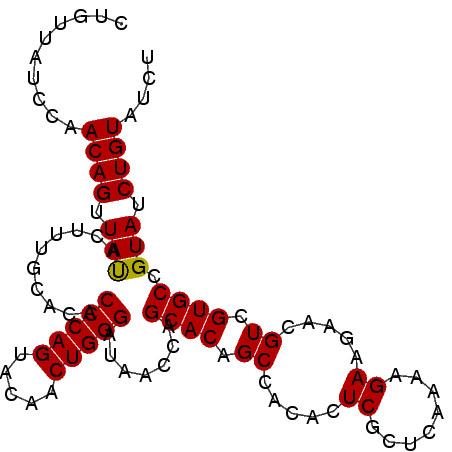
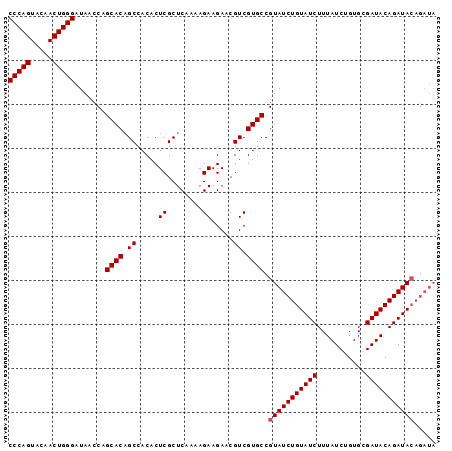
| Location | 14,045,818 – 14,045,917 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -34.64 |
| Energy contribution | -35.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.84 |
| SVM RNA-class probability | 0.999955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14045818 99 - 22407834 AGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCAGUGCUGGGUAUCCCAGUUGUACUGGGUGUGCAAAGUAUAACUGUUGUAUAAACG ..((((((...(.((((..(((((.......))))).)))).)(((((((..(((((((((.....))))).))))..)))...))))))))))..... ( -32.60) >DroSec_CAF1 134733 99 - 1 AGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUAUACUGGGUGUGCAAAGUAUAACUGUUGGAUAACAG .....(((.(((((((((.(((((.......)))))..))).)))))))))(((((((((((((((((..(....)..))))))))))..))))))).. ( -40.30) >DroSim_CAF1 137947 99 - 1 AGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUAUAACUGUUGGAUAACAG .....(((.(((((((((.(((((.......)))))..))).)))))))))(((((((((((((((((..(....)..))))))))))..))))))).. ( -40.30) >DroEre_CAF1 136761 99 - 1 AGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCCGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUGUAACUGUUGGAUAACAG ...........((((((..(((((.......)))))((.....))))))))(((((((((((((((((..(....)..))))))))))..))))))).. ( -36.20) >DroYak_CAF1 144257 99 - 1 AGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGACUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUGUAACUGUUGGAUAACAG .....(((.(((((((((.(((((.......)))))..))).)))))))))(((((((((((((((((..(....)..))))))))))..))))))).. ( -36.70) >consensus AGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUAUAACUGUUGGAUAACAG .....(((.(((((((((.(((((.......)))))..))).)))))))))(((((((((((((((((..(....)..))))))))))..))))))).. (-34.64 = -35.24 + 0.60)

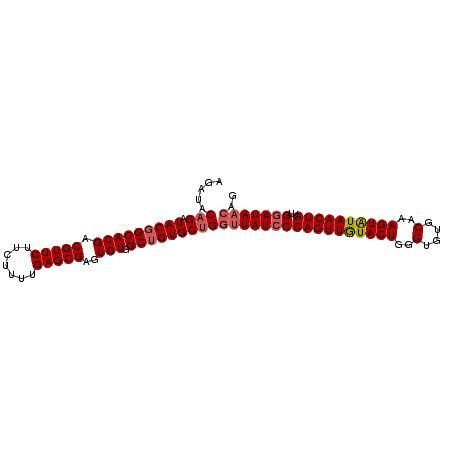
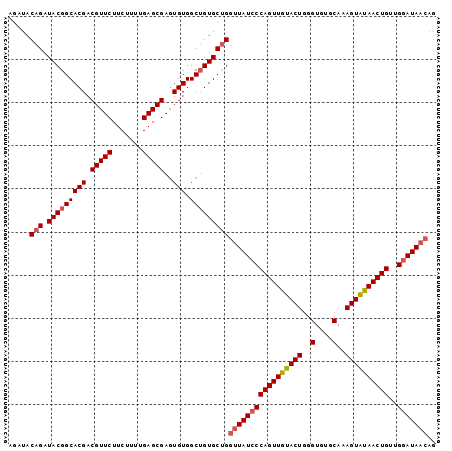
| Location | 14,045,846 – 14,045,943 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -27.62 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14045846 97 + 22407834 CCCAGUACAACUGGGAUACCCAGCACUGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCUUUAUCUGUGCGAUACAGAUACAGAUA (((((.....))))).......((((.((.....((........)).....)).))))..(((((((((((.((((.....)))).))))))))))) ( -30.80) >DroSec_CAF1 134761 97 + 1 CCCAGUAUAACUGGGAUAACCAGCACAGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCUUUAUCUGUGCGAUACAGAUACAGAUA (((((.....))))).......((((.((.....((........)).....)).))))..(((((((((((.((((.....)))).))))))))))) ( -30.80) >DroSim_CAF1 137975 97 + 1 CCCAGUACAACUGGGAUAACCAGCACAGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCUUUAUCUGUGCGAUACAGAUACAGAUA (((((.....))))).......((((.((.....((........)).....)).))))..(((((((((((.((((.....)))).))))))))))) ( -30.80) >DroEre_CAF1 136789 91 + 1 CCCAGUACAACUGGGAUAACCGGCACAGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCUUUAUCUGUGCGAUACAGAUA------ (((((.....))))).....((((((.((.....((........)).....)).))))))((((((((((...........))))))))))------ ( -30.20) >consensus CCCAGUACAACUGGGAUAACCAGCACAGCCACACUCGCUCAAAAGAAGAACGUCGUGCCGUAUCUGUAUCUUUAUCUGUGCGAUACAGAUACAGAUA (((((.....))))).......((((.((.....((........)).....)).)))).(((((((((((...........)))))))))))..... (-27.62 = -27.88 + 0.25)

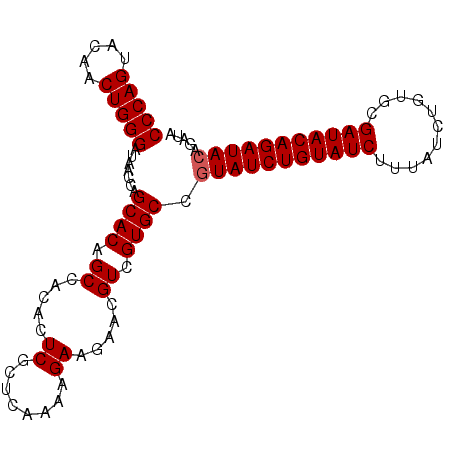
| Location | 14,045,846 – 14,045,943 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -35.31 |
| Energy contribution | -36.88 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.47 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14045846 97 - 22407834 UAUCUGUAUCUGUAUCGCACAGAUAAAGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCAGUGCUGGGUAUCCCAGUUGUACUGGG (((((((((((.((((.....)))).)))))))))))(.((((..(((((.......))))).)))).)((((((((((...))))))...)))).. ( -40.60) >DroSec_CAF1 134761 97 - 1 UAUCUGUAUCUGUAUCGCACAGAUAAAGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUAUACUGGG (((((((((((.((((.....)))).)))))))))))((((((..(((((.......)))))((.....)))))))).....(((((.....))))) ( -40.00) >DroSim_CAF1 137975 97 - 1 UAUCUGUAUCUGUAUCGCACAGAUAAAGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGG (((((((((((.((((.....)))).)))))))))))((((((..(((((.......)))))((.....)))))))).....(((((.....))))) ( -40.00) >DroEre_CAF1 136789 91 - 1 ------UAUCUGUAUCGCACAGAUAAAGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCCGGUUAUCCCAGUUGUACUGGG ------((((((((((...........))))))))))((((((..(((((.......)))))((.....)))))))).....(((((.....))))) ( -36.00) >consensus UAUCUGUAUCUGUAUCGCACAGAUAAAGAUACAGAUACGGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGG (((((((((((.((((.....)))).)))))))))))((((((..(((((.......)))))((.....)))))))).....(((((.....))))) (-35.31 = -36.88 + 1.56)

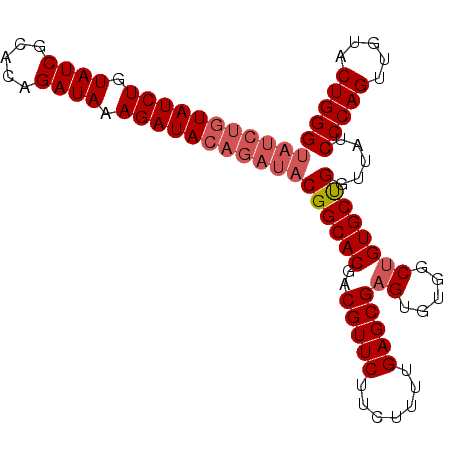
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:46 2006