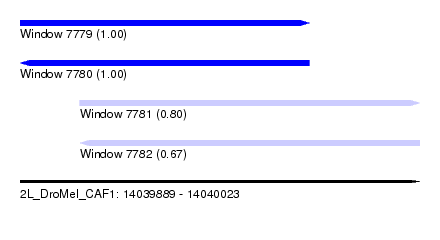
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,039,889 – 14,040,023 |
| Length | 134 |
| Max. P | 0.999919 |

| Location | 14,039,889 – 14,039,986 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -37.14 |
| Energy contribution | -36.66 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.26 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
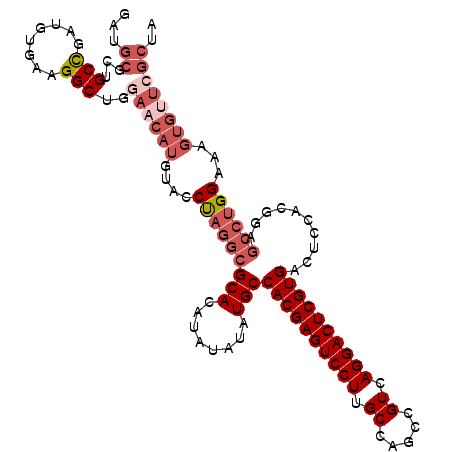
>2L_DroMel_CAF1 14039889 97 + 22407834 GACACACACACACUCGCACAUAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCC ..............((((((((..........(((((....).))))((((((((((((..(.....)..)))))))))))).....)))))))).. ( -37.30) >DroSec_CAF1 128836 97 + 1 GAGACCCACACACUCGCAUAUAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCC (((.........)))(((((((..........(((((....).))))((((((((((((..(.....)..)))))))))))).....)))))))... ( -35.40) >DroSim_CAF1 132033 97 + 1 GAGACCCACACACUCGCACAUAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCC (((.........)))(((((((..........(((((....).))))((((((((((((..(.....)..)))))))))))).....)))))))... ( -37.30) >DroEre_CAF1 130486 97 + 1 GAGUCCAGCACACUCGCACAUAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCC ((((.......))))(((((((..........(((((....).))))((((((((((((..(.....)..)))))))))))).....)))))))... ( -39.00) >DroYak_CAF1 138191 97 + 1 GAGUCGAACACACUUGCACAUAGCCAGCACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUGUGUGCGCC ...............(((((((..........(((((....).))))((((((((((((..(.....)..)))))))))))).....)))))))... ( -36.10) >consensus GAGACCCACACACUCGCACAUAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCC ..............((((((((..........(((((....).))))((((((((((((..(.....)..)))))))))))).....)))))))).. (-37.14 = -36.66 + -0.48)

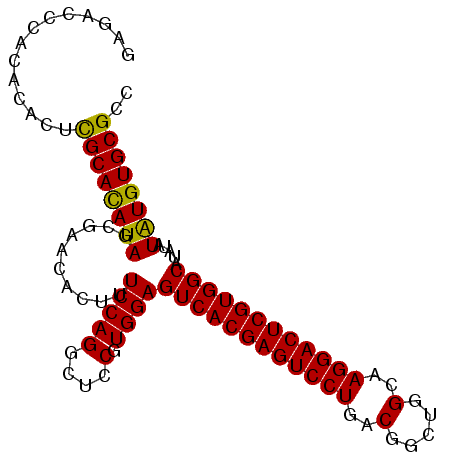
| Location | 14,039,889 – 14,039,986 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -41.08 |
| Consensus MFE | -37.18 |
| Energy contribution | -36.74 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14039889 97 - 22407834 GGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUAUGUGCGAGUGUGUGUGUGUC (((((((((((......((((((((((.((.....)).))))))))))((((((((((((((....)).))))....)))).))))))))))))))) ( -45.40) >DroSec_CAF1 128836 97 - 1 GGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUAUAUGCGAGUGUGUGGGUCUC (((.(((((........((((((((((.((.....)).))))))))))((((....((((((....)).))))((.....))))))))))).))).. ( -40.90) >DroSim_CAF1 132033 97 - 1 GGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUAUGUGCGAGUGUGUGGGUCUC (((.(((((........((((((((((.((.....)).))))))))))((((((((((((((....)).))))....)))).))))))))).))).. ( -42.00) >DroEre_CAF1 130486 97 - 1 GGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUAUGUGCGAGUGUGCUGGACUC (((((((.((((((.((((((((((((.((.....)).))))))))))........((((((....)).))))))))))))...)))))))...... ( -39.30) >DroYak_CAF1 138191 97 - 1 GGCGCACACAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGCUGGCUAUGUGCAAGUGUGUUCGACUC (((((((.((((((.((((((((((((.((.....)).)))))))))..........(((((....)).))))))))))))...)))))))...... ( -37.80) >consensus GGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUAUGUGCGAGUGUGUGGGUCUC .((((((.((((((.((((((((((((.((.....)).))))))))))........((((((....)).))))))))))))...))))))....... (-37.18 = -36.74 + -0.44)

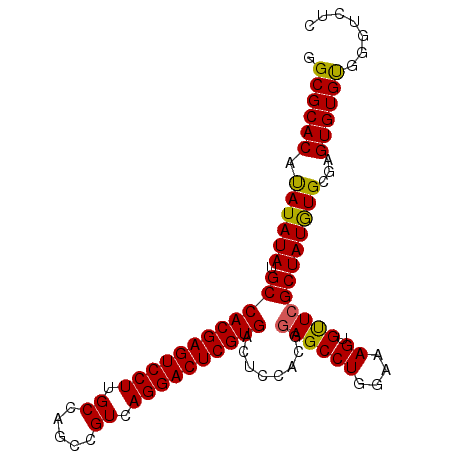
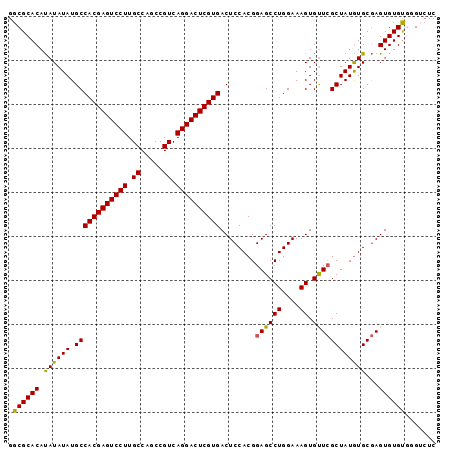
| Location | 14,039,909 – 14,040,023 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -41.48 |
| Consensus MFE | -33.84 |
| Energy contribution | -35.87 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14039909 114 + 22407834 UAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCCUAGGUACAUGUUCCAGCCUUCAAAUCGGCAGCGCAUC ..(((((((....((((((.(((((..((((((((((((..(.....)..)))))))))))).....)))).).)))).))....)))))..(((........)))...))... ( -44.30) >DroSec_CAF1 128856 114 + 1 UAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCCUAGGUACAUGUUCCAGCCUUCACAUCGGCAGCGCAUC ..(((((((....((((((.(((((..((((((((((((..(.....)..)))))))))))).....)))).).)))).))....)))))..(((........)))...))... ( -44.30) >DroSim_CAF1 132053 114 + 1 UAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCCUAGGUACAUGUUCCAGCCUUCACAUCGGCAGCGCAUC ..(((((((....((((((.(((((..((((((((((((..(.....)..)))))))))))).....)))).).)))).))....)))))..(((........)))...))... ( -44.30) >DroEre_CAF1 130506 114 + 1 UAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCCUAGGUACAUGUUCCAGCCUUCACAUCGGCAGCGCAUC ..(((((((....((((((.(((((..((((((((((((..(.....)..)))))))))))).....)))).).)))).))....)))))..(((........)))...))... ( -44.30) >DroYak_CAF1 138211 114 + 1 UAGCCAGCACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUGUGUGCGCCUAGGUACAUGUUCCAGCCUUCACAUCGGCAGCGCAUC ......((((..(((((....).))))((((((((((((..(.....)..))))))))))))......))))((((...((........)).(((........))).))))... ( -42.90) >DroPer_CAF1 171546 86 + 1 -----------UUCCAGGCACA--GGAGUCACGAGUCCUGACGACUCGCUAGGACUCGUG-------UAU--------GGUCACAUCUUCCAGCCUUGGCUCUAGCAAAAAAUC -----------..(((((((((--.(((((.((((((.....))))))....))))).))-------).(--------((.........)))))).)))............... ( -28.80) >consensus UAGCGAACACUUUCCAGGCUCCGUGGAGUCACGAGUCCUGACGGCUGGCAAGGACUCGUGGCAUAUAUAUGUGCGCCUAGGUACAUGUUCCAGCCUUCACAUCGGCAGCGCAUC ..(((..........(((((....(((((((((((((((..(.....)..))))))))))))....((((((((......))))))))))))))))............)))... (-33.84 = -35.87 + 2.03)

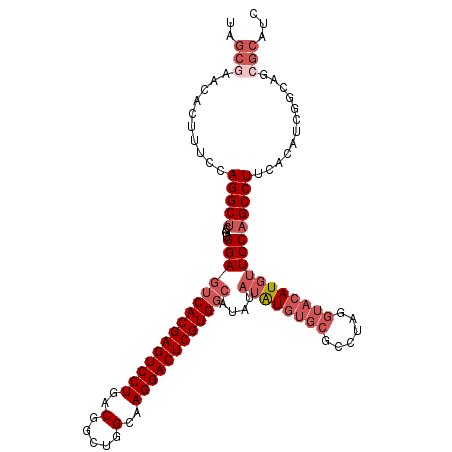
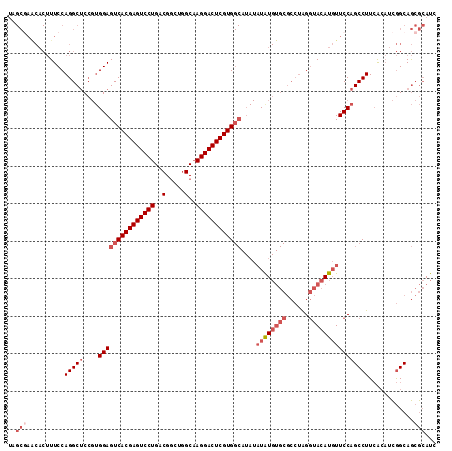
| Location | 14,039,909 – 14,040,023 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -41.43 |
| Consensus MFE | -33.65 |
| Energy contribution | -35.83 |
| Covariance contribution | 2.18 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14039909 114 - 22407834 GAUGCGCUGCCGAUUUGAAGGCUGGAACAUGUACCUAGGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUA ...((...(((........)))..((((((....(((((((((........)))((((((((((.((.....)).))))))))))..........))))))...)))))))).. ( -43.90) >DroSec_CAF1 128856 114 - 1 GAUGCGCUGCCGAUGUGAAGGCUGGAACAUGUACCUAGGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUA ...((...(((........)))..((((((....(((((((((........)))((((((((((.((.....)).))))))))))..........))))))...)))))))).. ( -43.90) >DroSim_CAF1 132053 114 - 1 GAUGCGCUGCCGAUGUGAAGGCUGGAACAUGUACCUAGGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUA ...((...(((........)))..((((((....(((((((((........)))((((((((((.((.....)).))))))))))..........))))))...)))))))).. ( -43.90) >DroEre_CAF1 130506 114 - 1 GAUGCGCUGCCGAUGUGAAGGCUGGAACAUGUACCUAGGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUA ...((...(((........)))..((((((....(((((((((........)))((((((((((.((.....)).))))))))))..........))))))...)))))))).. ( -43.90) >DroYak_CAF1 138211 114 - 1 GAUGCGCUGCCGAUGUGAAGGCUGGAACAUGUACCUAGGCGCACACAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGCUGGCUA ...((((((((........)))............(((((((((........)))((((((((((.((.....)).))))))))))..........))))))..)))))...... ( -41.40) >DroPer_CAF1 171546 86 - 1 GAUUUUUUGCUAGAGCCAAGGCUGGAAGAUGUGACC--------AUA-------CACGAGUCCUAGCGAGUCGUCAGGACUCGUGACUCC--UGUGCCUGGAA----------- ...............(((.(((.(((..(((....)--------)).-------((((((((((..((...))..))))))))))..)))--...))))))..----------- ( -31.60) >consensus GAUGCGCUGCCGAUGUGAAGGCUGGAACAUGUACCUAGGCGCACAUAUAUAUGCCACGAGUCCUUGCCAGCCGUCAGGACUCGUGACUCCACGGAGCCUGGAAAGUGUUCGCUA ...((...(((........)))..((((((....(((((((((........)))((((((((((.((.....)).))))))))))..........))))))...)))))))).. (-33.65 = -35.83 + 2.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:41 2006