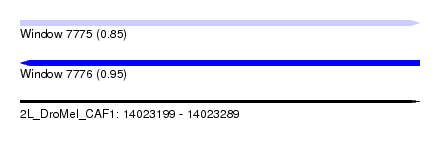
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,023,199 – 14,023,289 |
| Length | 90 |
| Max. P | 0.954566 |

| Location | 14,023,199 – 14,023,289 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -11.20 |
| Energy contribution | -12.55 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
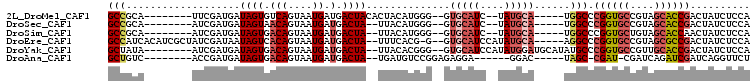
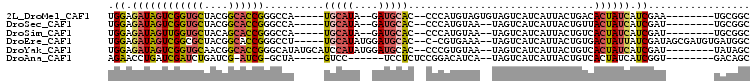
>2L_DroMel_CAF1 14023199 90 + 22407834 GCCGCA--------UUCGAUGAUAGUGUCAGUAAUGAUGACUACACUACAUGGG--GUGCAUC--UAUGCA-----UGGCCCGGUGCCGUAGCACCGACUAUCUCCA ...(((--------(((.(((.((((((.(((.......)))))))))))).))--))))...--.....(-----(((..((((((....)))))).))))..... ( -31.10) >DroSec_CAF1 110743 88 + 1 GCCGCA--------AUCGAUGAUAGUAACAGUAAUGAUGACUA--UUACAUGGG--GUGCAUC--UAUGCA-----UGGCCCGGUGCCGUAGCACCGACUAUCUCCA ......--------...((.((((((....((((((.....))--))))..(((--(((((..--..))))-----)..)))(((((....))))).)))))))).. ( -26.90) >DroSim_CAF1 115126 88 + 1 GCCGCA--------AUCGAUGAUAGUGACAGUAAUGAUGACUA--UUACAUGGG--GUGCAUC--UAUGCA-----UGGCCCGGUGCUGUAGCACCAACUAUCUCCA ......--------...((.((((((....((((((.....))--))))..(((--(((((..--..))))-----)..)))(((((....))))).)))))))).. ( -27.50) >DroEre_CAF1 113815 97 + 1 GCCAUCACAUCGCUAUCGAUAAUAGUCACAGUAAUGAUGACUA--UUUCACG-G--GUGCAUCCAUAUGCA-----AGGCCCGGUGCCGUAGCGCCGACUAUCUCCA (((............(((..((((((((((....)).))))))--))...))-)--.(((((....)))))-----.))).((((((....)))))).......... ( -30.00) >DroYak_CAF1 121161 95 + 1 GCUAUA--------AUCGAUGAUAGUGACAGUAAUGAUGACUA--UUACACGGG--GUGCAUCCAUAUGGAUGCAUAUGCCCGGUGCCGUUGCACCGACUAUCUCCA ......--------...((.((((((....((((((.....))--))))..(((--((((((((....))))))))...)))(((((....))))).)))))))).. ( -34.90) >DroAna_CAF1 110130 84 + 1 GCUGUC--------ACCGAUGAUAGUGACAGUAAUGAUGACUA--UGAUGUCCGGAGAGGA------GGAC-----UAGC-CGAU-CGAUCAGAUCGAUCAGGUUCU ((((((--------((........))))))))...........--....((((........------))))-----.(((-((((-((((...))))))).)))).. ( -27.30) >consensus GCCGCA________AUCGAUGAUAGUGACAGUAAUGAUGACUA__UUACAUGGG__GUGCAUC__UAUGCA_____UGGCCCGGUGCCGUAGCACCGACUAUCUCCA .................((.((((((...............................(((((....)))))..........((((((....)))))))))))))).. (-11.20 = -12.55 + 1.35)
| Location | 14,023,199 – 14,023,289 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -15.41 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
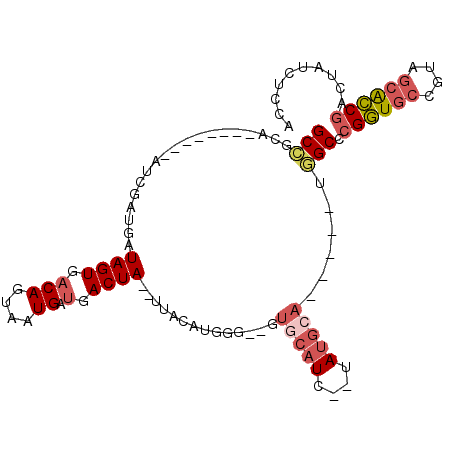
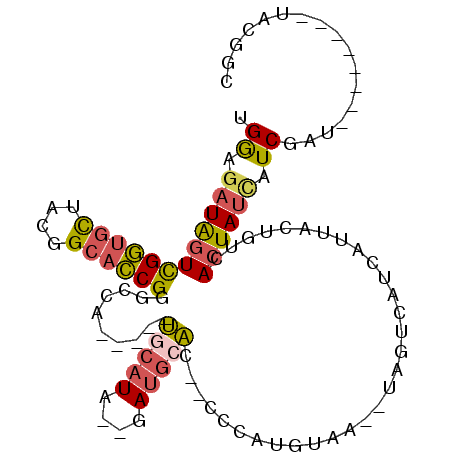
>2L_DroMel_CAF1 14023199 90 - 22407834 UGGAGAUAGUCGGUGCUACGGCACCGGGCCA-----UGCAUA--GAUGCAC--CCCAUGUAGUGUAGUCAUCAUUACUGACACUAUCAUCGAA--------UGCGGC .........(((((((....)))))))(((.-----.((((.--((((...--.....((((((((((.......))).)))))))))))..)--------)))))) ( -30.40) >DroSec_CAF1 110743 88 - 1 UGGAGAUAGUCGGUGCUACGGCACCGGGCCA-----UGCAUA--GAUGCAC--CCCAUGUAA--UAGUCAUCAUUACUGUUACUAUCAUCGAU--------UGCGGC .........(((((((....)))))))(((.-----.(((..--((((...--.....((((--((((.......))))))))...))))...--------)))))) ( -28.70) >DroSim_CAF1 115126 88 - 1 UGGAGAUAGUUGGUGCUACAGCACCGGGCCA-----UGCAUA--GAUGCAC--CCCAUGUAA--UAGUCAUCAUUACUGUCACUAUCAUCGAU--------UGCGGC ..((((((((.(((((....)))))(((...-----((((..--..)))).--)))..((((--(.......)))))....)))))).))...--------...... ( -23.80) >DroEre_CAF1 113815 97 - 1 UGGAGAUAGUCGGCGCUACGGCACCGGGCCU-----UGCAUAUGGAUGCAC--C-CGUGAAA--UAGUCAUCAUUACUGUGACUAUUAUCGAUAGCGAUGUGAUGGC .........((((.((....)).))))((((-----..(((((((......--)-)))((((--(((((((.......))))))))).)).......)))..).))) ( -29.70) >DroYak_CAF1 121161 95 - 1 UGGAGAUAGUCGGUGCAACGGCACCGGGCAUAUGCAUCCAUAUGGAUGCAC--CCCGUGUAA--UAGUCAUCAUUACUGUCACUAUCAUCGAU--------UAUAGC ..((((((((((((((....))))))((((..(((((((....))))))).--.....((((--(.......))))))))))))))).))...--------...... ( -32.80) >DroAna_CAF1 110130 84 - 1 AGAACCUGAUCGAUCUGAUCG-AUCG-GCUA-----GUCC------UCCUCUCCGGACAUCA--UAGUCAUCAUUACUGUCACUAUCAUCGGU--------GACAGC .((....(((((((...))))-)))(-((((-----((((------........))))....--))))).))....((((((((......)))--------))))). ( -27.60) >consensus UGGAGAUAGUCGGUGCUACGGCACCGGGCCA_____UGCAUA__GAUGCAC__CCCAUGUAA__UAGUCAUCAUUACUGUCACUAUCAUCGAU________UACGGC .((.((((((((((((....))))))..........(((((....)))))...............................)))))).))................. (-15.41 = -15.73 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:35 2006