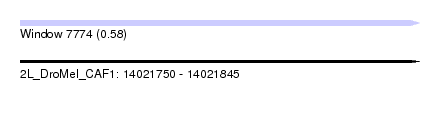
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,021,750 – 14,021,845 |
| Length | 95 |
| Max. P | 0.580503 |

| Location | 14,021,750 – 14,021,845 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -19.18 |
| Energy contribution | -20.24 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
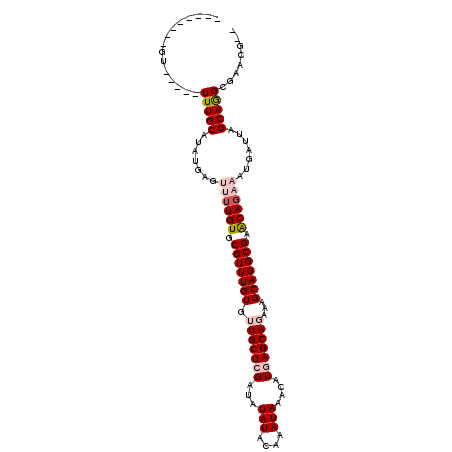
>2L_DroMel_CAF1 14021750 95 + 22407834 --------GU-----UUUGCAUAUGAGCUUUGUGCGUUUGUGUUGCUCGAUAUAUACAAAUAAACAUAAGCAGCCAGCAGGCGAGCAGAAACGAUUAGCAGGCCGAAA-- --------.(-----(((((......(((((((..((((((((..........))))))))..))).)))).(((....)))..))))))..................-- ( -22.80) >DroVir_CAF1 201447 95 + 1 --------UU-----UUUGCAUAUGUGUUUUGUGCGUUUGUGUUGCUCGAUAUAUACAAAUAAACAUGAGCAGAAAGCAGGCGCACAGCAAUGAUUAGCAAGCGUACG-- --------..-----(((((.....((((((((((((((((.(((((((...(((....)))....)))))))...)))))))))))).))))....)))))......-- ( -28.50) >DroGri_CAF1 128793 94 + 1 ---------U-----UUUGCAUAUGAGUUUUGUGCGUUUGUGUUGCUCGAUAUAUACAAAUAAACAUGAGCACAAAGCAGGCGCACAGAAAUGAUUAGCAAGCGAACG-- ---------.-----(((((..((.(.((((((((((((((..((((((...(((....)))....))))))....)))))))))))))).).))..)))))......-- ( -29.30) >DroWil_CAF1 156436 97 + 1 --------GU-----UUUGCAUAUGAAUAUUGUGCGUUUGUGUUGCUCGAUAUAUACAAAUAAACAUGAGCAGAAAGCAGGCGAACAGAAGUGAUUAGCAGGCAAAGGCA --------.(-----(((((.........((((.(((((((.(((((((...(((....)))....)))))))...))))))).))))..((.....))..))))))... ( -23.40) >DroMoj_CAF1 169109 103 + 1 UUAUUUCAUU-----UUUGCAUAUGUGUUUUGUGCGUUUGUGUUGCUCGAUAUAUACAAAUAAACAUGAGCACAAAGCAGGCGCACAGAAAUGAUUAGCAGGCGCACG-- ..........-----(((((.......((((((((((((((..((((((...(((....)))....))))))....)))))))))))))).......)))))......-- ( -28.84) >DroAna_CAF1 108778 100 + 1 --------GUAUUGCUUUGCAUAUGGGCUGUGUGCGUUUGUGUUGCUCGAUAUAUACAAAUAAACAUAAGCAGCCAGCAGGCGAGCAGAAGCGAUUAGCAGGCCGAGC-- --------((((((((((((......(((((((..((((((((..........))))))))..)))).))).(((....)))..)))).))))))..)).........-- ( -27.70) >consensus ________GU_____UUUGCAUAUGAGUUUUGUGCGUUUGUGUUGCUCGAUAUAUACAAAUAAACAUGAGCAGAAAGCAGGCGAACAGAAAUGAUUAGCAGGCGAACG__ ...............(((((.......((((((.(((((((.(((((((...(((....)))....)))))))...))))))).)))))).......)))))........ (-19.18 = -20.24 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:33 2006