
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,978,137 – 13,978,293 |
| Length | 156 |
| Max. P | 0.993136 |

| Location | 13,978,137 – 13,978,253 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.91 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
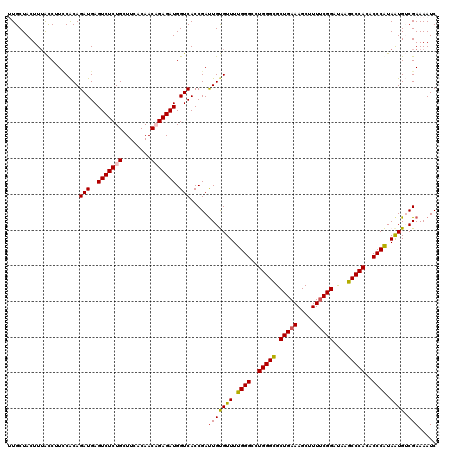
>2L_DroMel_CAF1 13978137 116 - 22407834 UCAGCGCCCAGACCCAAAACACAAUCGGUGACCAUCUCUGUUGUGAAGCAGAGACUCAUCUGUGGACGAUAAAGUAGCAAAAAAAAAAAAAAAACAGGUUUGGUUUCAUAGAAUAC .......(((((((.....((((...(((((...((((((((....)))))))).))))))))).((......)).....................)))))))............. ( -27.50) >DroSec_CAF1 65641 106 - 1 UCAGCGCCCAGGCCCAAAACACAAUCGGUGACCAUCUCUGUUGUGAAGCAGAGACUCAUCUGUGGAAGGUAAAGUAGCAAA----------AAACAGGUUUGGUUUCAUAUAAUAC .......(((((((.....((((...(((((...((((((((....)))))))).)))))))))....((......))...----------.....)))))))............. ( -26.70) >DroSim_CAF1 69416 106 - 1 UCAGCGCCCAGGCCCAAAACACAAUCGGUGACCAUCUCUGUUGUGAAGCAGAGACUCAUCUGUGGAAGGUAAAGUAGCAAA----------AAACAGGUUUGGUUUCAUAUAAUAC .......(((((((.....((((...(((((...((((((((....)))))))).)))))))))....((......))...----------.....)))))))............. ( -26.70) >DroEre_CAF1 68442 105 - 1 UCAGCACCCAGGCCCAAAACGCAGUCGGUGACCAUCUCAGUUGUAAAGCAGAGACUCAUCUGCGAA-GGCCAAGUAGCACA----------AAACGGGUUUGGGUUCAUAUAAUGC ...(.((((((((((....(((((...((((...((((.(((....))).)))).)))))))))..-.((......))...----------....)))))))))).)......... ( -34.40) >DroYak_CAF1 74959 106 - 1 UCAGCACCCAGGCCCAAAACGCAAUCGGUGACCAUCUCGGUUGUAAAGCAGAGACUCAUCUGUGUAUGGCAAAGUAGCACG----------AAACAGGUUUGUAUUCAUAUAAUAU ...((((....(((....(((((...(((((...((((.(((....))).)))).))))))))))..)))...)).))(((----------((.....)))))............. ( -21.40) >consensus UCAGCGCCCAGGCCCAAAACACAAUCGGUGACCAUCUCUGUUGUGAAGCAGAGACUCAUCUGUGGAAGGUAAAGUAGCAAA__________AAACAGGUUUGGUUUCAUAUAAUAC .......(((((((.....((((...(((((...((((((((....)))))))).)))))))))....((......))..................)))))))............. (-24.04 = -24.04 + -0.00)


| Location | 13,978,173 – 13,978,293 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.04 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13978173 120 + 22407834 UUGCUACUUUAUCGUCCACAGAUGAGUCUCUGCUUCACAACAGAGAUGGUCACCGAUUGUGUUUUGGGUCUGGGCGCUGAAAGCUUUUCGGAUAAGCCCACACCCAUAAUGUCGAAAAUC ..(((......(((((....)))))(((((((........))))))))))...((((...(((.(((((.(((((.(((((.....)))))....))))).))))).)))))))...... ( -38.20) >DroSec_CAF1 65667 120 + 1 UUGCUACUUUACCUUCCACAGAUGAGUCUCUGCUUCACAACAGAGAUGGUCACCGAUUGUGUUUUGGGCCUGGGCGCUGAAAGCUUUUCGGAUAAGCCCACACCCAUAAUGUCGAAAAUC ....................(((..(((((((........))))))).)))..((((...(((.((((..(((((.(((((.....)))))....)))))..)))).)))))))...... ( -35.20) >DroSim_CAF1 69442 120 + 1 UUGCUACUUUACCUUCCACAGAUGAGUCUCUGCUUCACAACAGAGAUGGUCACCGAUUGUGUUUUGGGCCUGGGCGCUGAAAGCUUUUCGGAUAAGCCCACACCCAUAAUGUCGAAAAUC ....................(((..(((((((........))))))).)))..((((...(((.((((..(((((.(((((.....)))))....)))))..)))).)))))))...... ( -35.20) >DroEre_CAF1 68468 119 + 1 GUGCUACUUGGCC-UUCGCAGAUGAGUCUCUGCUUUACAACUGAGAUGGUCACCGACUGCGUUUUGGGCCUGGGUGCUGAAAGCUUUACGGAUAAGCCCACACCCGUAAUGGCGAAAAUC .(((((...((((-(.(((((....(((((............)))))((...))..)))))....))))).(((((.((...((((.......)))).)))))))....)))))...... ( -36.90) >DroYak_CAF1 74985 120 + 1 GUGCUACUUUGCCAUACACAGAUGAGUCUCUGCUUUACAACCGAGAUGGUCACCGAUUGCGUUUUGGGCCUGGGUGCUGAAAGCUUUUCGGACAAGCCCACACCCGUAGUGGCGAAAAUC .......((((((((((...(((..(((((.(........).))))).)))..............(((..(((((.(((((.....)))))....)))))..))))).)))))))).... ( -35.40) >consensus UUGCUACUUUACCUUCCACAGAUGAGUCUCUGCUUCACAACAGAGAUGGUCACCGAUUGUGUUUUGGGCCUGGGCGCUGAAAGCUUUUCGGAUAAGCCCACACCCAUAAUGUCGAAAAUC ....................(((..(((((((........))))))).))).....(((((((.((((..(((((.(((((.....)))))....)))))..)))).)))).)))..... (-30.12 = -30.04 + -0.08)


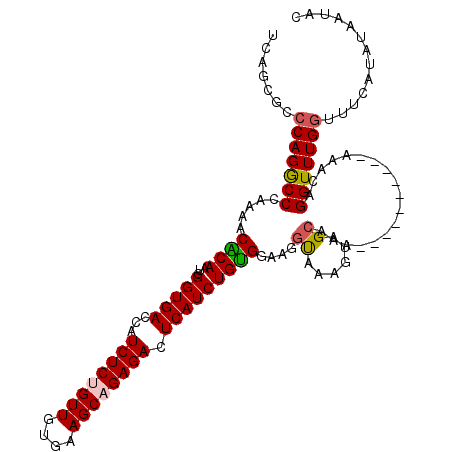
| Location | 13,978,173 – 13,978,293 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -34.06 |
| Energy contribution | -33.22 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13978173 120 - 22407834 GAUUUUCGACAUUAUGGGUGUGGGCUUAUCCGAAAAGCUUUCAGCGCCCAGACCCAAAACACAAUCGGUGACCAUCUCUGUUGUGAAGCAGAGACUCAUCUGUGGACGAUAAAGUAGCAA .....(((......(((((.(((((((....))...((.....))))))).)))))...((((...(((((...((((((((....)))))))).)))))))))..)))........... ( -38.60) >DroSec_CAF1 65667 120 - 1 GAUUUUCGACAUUAUGGGUGUGGGCUUAUCCGAAAAGCUUUCAGCGCCCAGGCCCAAAACACAAUCGGUGACCAUCUCUGUUGUGAAGCAGAGACUCAUCUGUGGAAGGUAAAGUAGCAA .((((((.(((...(((((.(((((((....))...((.....))))))).)))))..........(((((...((((((((....)))))))).)))))))).)))))).......... ( -41.10) >DroSim_CAF1 69442 120 - 1 GAUUUUCGACAUUAUGGGUGUGGGCUUAUCCGAAAAGCUUUCAGCGCCCAGGCCCAAAACACAAUCGGUGACCAUCUCUGUUGUGAAGCAGAGACUCAUCUGUGGAAGGUAAAGUAGCAA .((((((.(((...(((((.(((((((....))...((.....))))))).)))))..........(((((...((((((((....)))))))).)))))))).)))))).......... ( -41.10) >DroEre_CAF1 68468 119 - 1 GAUUUUCGCCAUUACGGGUGUGGGCUUAUCCGUAAAGCUUUCAGCACCCAGGCCCAAAACGCAGUCGGUGACCAUCUCAGUUGUAAAGCAGAGACUCAUCUGCGAA-GGCCAAGUAGCAC .......((...(((((((((.(((((.......)))))....)))))).((((.....(((((...((((...((((.(((....))).)))).)))))))))..-))))..))))).. ( -38.40) >DroYak_CAF1 74985 120 - 1 GAUUUUCGCCACUACGGGUGUGGGCUUGUCCGAAAAGCUUUCAGCACCCAGGCCCAAAACGCAAUCGGUGACCAUCUCGGUUGUAAAGCAGAGACUCAUCUGUGUAUGGCAAAGUAGCAC .(((((.((((.(((((((.((((..(((..(((.....))).))))))).)))).....((((((((.(.....)))))))))...(((((......)))))))))))))))))..... ( -38.90) >consensus GAUUUUCGACAUUAUGGGUGUGGGCUUAUCCGAAAAGCUUUCAGCGCCCAGGCCCAAAACACAAUCGGUGACCAUCUCUGUUGUGAAGCAGAGACUCAUCUGUGGAAGGUAAAGUAGCAA ...........((((((((((.(((((.......)))))....))))))..(((.....((((...(((((...((((((((....)))))))).)))))))))...)))...))))... (-34.06 = -33.22 + -0.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:22 2006