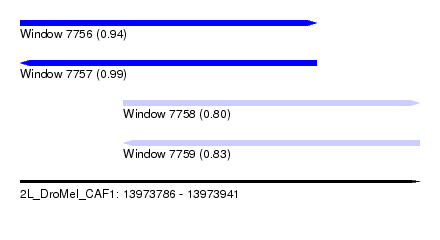
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,973,786 – 13,973,941 |
| Length | 155 |
| Max. P | 0.992449 |

| Location | 13,973,786 – 13,973,901 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13973786 115 + 22407834 UUAUCUAUUGGAUUGAUUAACCACACAGAAACUCUGUAUGGGAAUAGAUACAUAUAUGACAAGUACUGAAAAAGGAAACGCUGAUACGACUGGUUGGUAUCAGCGUUUUCUUUUC .((((((((...........(((.((((.....)))).))).))))))))...................((((((((((((((((((.(.....).)))))))))))))))))). ( -35.80) >DroSec_CAF1 61170 96 + 1 -----------AAUGAUUUAGCACUCAGAAACUCUGUAUGGGAAUAGGU----AUAUGAC----ACUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUC -----------.....(((((...(((...((.((((......))))))----...))).----.)))))((((((((.((((((((.(.....).)))))))).)))))))).. ( -23.80) >DroSim_CAF1 64947 96 + 1 -----------AAUGAUUUAGCACUCAGAAACUCUGUAUGGGAAUAGGU----AUAUGAC----ACUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUC -----------.....(((((...(((...((.((((......))))))----...))).----.)))))((((((((.((((((((.(.....).)))))))).)))))))).. ( -23.80) >consensus ___________AAUGAUUUAGCACUCAGAAACUCUGUAUGGGAAUAGGU____AUAUGAC____ACUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUC .....................((..(((.....)))..)).............................(((((((((.((((((((.(.....).)))))))).))))))))). (-21.40 = -21.40 + 0.00)

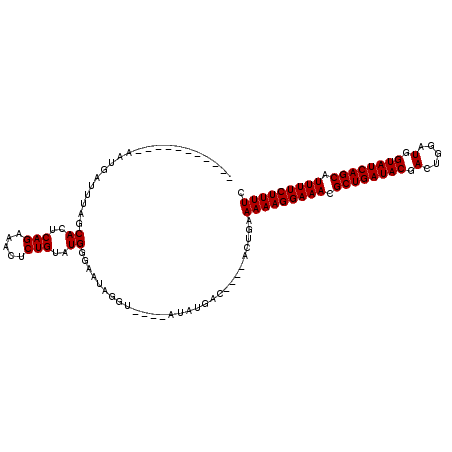
| Location | 13,973,786 – 13,973,901 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13973786 115 - 22407834 GAAAAGAAAACGCUGAUACCAACCAGUCGUAUCAGCGUUUCCUUUUUCAGUACUUGUCAUAUAUGUAUCUAUUCCCAUACAGAGUUUCUGUGUGGUUAAUCAAUCCAAUAGAUAA ((((((.((((((((((((.........)))))))))))).))))))..................((((((((.((((((((.....))))))))...........)))))))). ( -35.10) >DroSec_CAF1 61170 96 - 1 GAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAGU----GUCAUAU----ACCUAUUCCCAUACAGAGUUUCUGAGUGCUAAAUCAUU----------- ((((((.((((((((((((.........)))))))))))).))))))....----.......----.........(((.(((.....))).)))..........----------- ( -22.10) >DroSim_CAF1 64947 96 - 1 GAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAGU----GUCAUAU----ACCUAUUCCCAUACAGAGUUUCUGAGUGCUAAAUCAUU----------- ((((((.((((((((((((.........)))))))))))).))))))....----.......----.........(((.(((.....))).)))..........----------- ( -22.10) >consensus GAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAGU____GUCAUAU____ACCUAUUCCCAUACAGAGUUUCUGAGUGCUAAAUCAUU___________ ((((((.((((((((((((.........)))))))))))).))))))............................(((.(((.....))).)))..................... (-23.22 = -23.00 + -0.22)

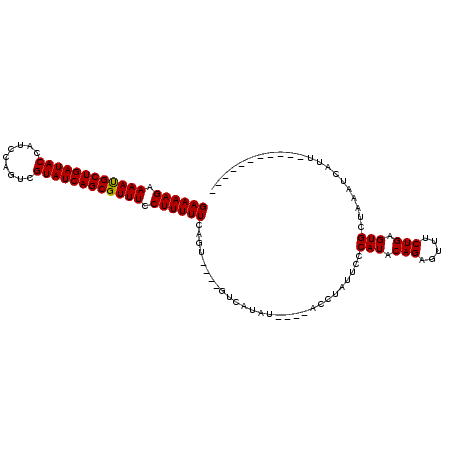
| Location | 13,973,826 – 13,973,941 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.46 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13973826 115 + 22407834 GGAAUAGAUACAUAUAUGACAAGUACUGAAAAAGGAAACGCUGAUACGACUGGUUGGUAUCAGCGUUUUCUUUUCCAGUACUCGUAAAGUAUUCGUAUAAAUCUUCUUAACUAUU ((((.(.((((..((((.((.(((((((.((((((((((((((((((.(.....).)))))))))))))))))).))))))).))...))))..))))...).))))........ ( -42.60) >DroSec_CAF1 61199 107 + 1 GGAAUAGGU----AUAUGAC----ACUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUCCAGUAGUCGUAAAGUAUUCGUAAAAAUCUUCUUAACCAUA .(((((..(----.((((((----((((.(((((((((.((((((((.(.....).)))))))).))))))))).)))).)))))))..)))))..................... ( -34.40) >DroSim_CAF1 64976 107 + 1 GGAAUAGGU----AUAUGAC----ACUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUCCAGUAGUCGUAAAGUAUUCGUAAAAAUCUUCUUAACCAUA .(((((..(----.((((((----((((.(((((((((.((((((((.(.....).)))))))).))))))))).)))).)))))))..)))))..................... ( -34.40) >consensus GGAAUAGGU____AUAUGAC____ACUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUCCAGUAGUCGUAAAGUAUUCGUAAAAAUCUUCUUAACCAUA .(((((..(.....((((((....((((.(((((((((.((((((((.(.....).)))))))).))))))))).)))).)))))))..)))))..................... (-28.70 = -29.03 + 0.33)


| Location | 13,973,826 – 13,973,941 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -31.21 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -6.19 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13973826 115 - 22407834 AAUAGUUAAGAAGAUUUAUACGAAUACUUUACGAGUACUGGAAAAGAAAACGCUGAUACCAACCAGUCGUAUCAGCGUUUCCUUUUUCAGUACUUGUCAUAUAUGUAUCUAUUCC ............(((..(((((.(((....((((((((((((((((.((((((((((((.........)))))))))))).))))))))))))))))..))).)))))..))).. ( -41.90) >DroSec_CAF1 61199 107 - 1 UAUGGUUAAGAAGAUUUUUACGAAUACUUUACGACUACUGGAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAGU----GUCAUAU----ACCUAUUCC ...(((...((((((((....)))).))))..(((.((((((((((.((((((((((((.........)))))))))))).))))))))))----)))....----)))...... ( -34.60) >DroSim_CAF1 64976 107 - 1 UAUGGUUAAGAAGAUUUUUACGAAUACUUUACGACUACUGGAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAGU----GUCAUAU----ACCUAUUCC ...(((...((((((((....)))).))))..(((.((((((((((.((((((((((((.........)))))))))))).))))))))))----)))....----)))...... ( -34.60) >consensus UAUGGUUAAGAAGAUUUUUACGAAUACUUUACGACUACUGGAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAGU____GUCAUAU____ACCUAUUCC .((((....((((((((....)))).))))..(((.((((((((((.((((((((((((.........)))))))))))).))))))))))....)))..........))))... (-31.21 = -31.10 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:19 2006