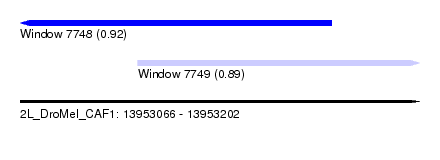
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,953,066 – 13,953,202 |
| Length | 136 |
| Max. P | 0.919691 |

| Location | 13,953,066 – 13,953,172 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -23.20 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
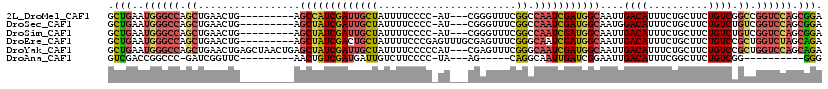
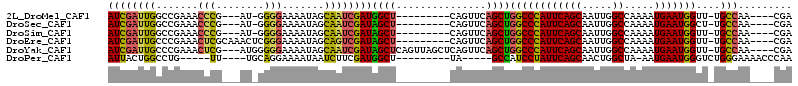
>2L_DroMel_CAF1 13953066 106 - 22407834 GCUGAAUGGGCCAGCUGAACUG---------AGCCAUCGAUUGCUAUUUUCCCC-AU---CGGGUUUCGGCCAAUCGAUGGCAAUUGACAUUUCUGCUUCUGUCGGCCGGUCCAGCGGA ((((.((.((((..........---------.(((((((((((((.....(((.-..---.)))....)).)))))))))))....((((..........)))))))).)).))))... ( -44.00) >DroSec_CAF1 40872 106 - 1 GCUGAAUGGGCCAGCUGAACUG---------AGCUAUCGAUUGCUAUUUUCCCC-AU---CGGGUUUCGGCCAAUCGAUGGCAAUGGACAUUUCUGCUUCUGUCUGUCGGUCCAGCGGA ..........((.((((.((((---------((((((((((((((.....(((.-..---.)))....)).)))))))))))...(((((..........))))).))))).)))))). ( -38.20) >DroSim_CAF1 41073 106 - 1 GCUGAAUGGGCCAGCUGAACUG---------AGCUAUCGAUUGCUAUUUUCCCC-AU---CGGGUUUCGGCCAAUCGAUGGCAAUUGACAUUUCUGCUUCUGUCUGUCGGUCCAGCGGA ..........((.((((.((((---------((((((((((((((.....(((.-..---.)))....)).)))))))))))....((((..........))))..))))).)))))). ( -36.90) >DroEre_CAF1 41955 110 - 1 GCUGAAUGGGCCAGCUGAACUG---------AGCUAUCGACUGCUAUUUUCCCGAGUUUGCGAGUUUCGGGCAAUCGAUGGCAAUUGACAUUUCUGCUUCUGUCCGCUGGUCUAGCAGA .(((..(((((((((.((...(---------(((..((((.(((((((((((((((.........)))))).))..))))))).)))).......))))...)).))))))))).))). ( -39.40) >DroYak_CAF1 42299 116 - 1 GCUGAAUGGGCCAGCUGAACUGAGCUAACUGAGCUAUCGAUUGCUAUUUUCCCCCAU---CGAGUUUCGGGCAAUCGAUGGCAAUUGACAUUUCUGCUUCUGUCCGCUGGUCCAGCAGA .(((..(((((((((.((...((((.......(((((((((((((...(((......---.))).....)))))))))))))....((....)).))))...)).))))))))).))). ( -46.00) >DroAna_CAF1 40401 90 - 1 GUCGACCGGCCC-GAUCGGUUC---------AACUGUCGAUGAUUGUCUUCCCC-UA---AG-----CAGGCAAUUGAUCGGAAUUGACAUUUCGGCUUCUGUCGG----------GGG ((((((((..((-((.(((...---------..))))))..((((((((.(...-..---.)-----.)))))))))..)))..))))).(..((((....)))).----------.). ( -26.90) >consensus GCUGAAUGGGCCAGCUGAACUG_________AGCUAUCGAUUGCUAUUUUCCCC_AU___CGGGUUUCGGCCAAUCGAUGGCAAUUGACAUUUCUGCUUCUGUCCGCCGGUCCAGCGGA .(((..((((((.((.................(((((((((((((.......................)).)))))))))))....((((..........)))).)).)))))).))). (-23.20 = -24.10 + 0.90)
| Location | 13,953,106 – 13,953,202 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -20.01 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
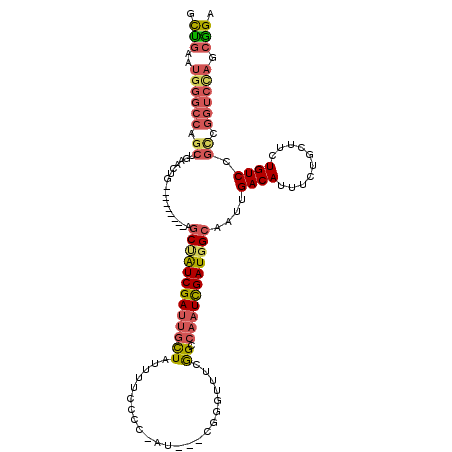
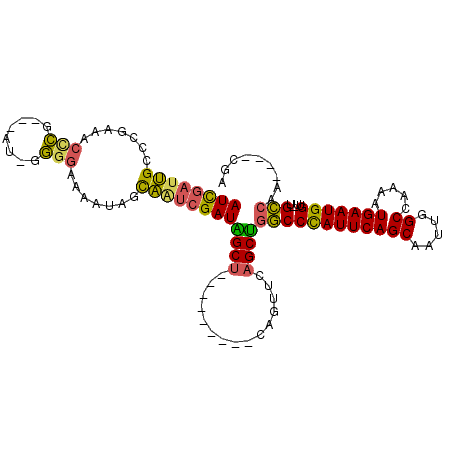
>2L_DroMel_CAF1 13953106 96 + 22407834 AUCGAUUGGCCGAAACCCG---AU-GGGGAAAAUAGCAAUCGAUGGCU---------CAGUUCAGCUGGCCCAUUCAGCAAUUGGCCAAAAUGAAUGGUU-UGCCAA----CGA .(((.((((((((..(((.---..-.)))......((....(((((..---------(((.....)))..)))))..))..))))))))......(((..-..))).----))) ( -31.40) >DroSec_CAF1 40912 96 + 1 AUCGAUUGGCCGAAACCCG---AU-GGGGAAAAUAGCAAUCGAUAGCU---------CAGUUCAGCUGGCCCAUUCAGCAAUUGGCCAAAAUGAAUGGCU-UGCCAA----CGA .(((.((((((((..(((.---..-)))(((...(((........)))---------...))).(((((.....)))))..))))))))......(((..-..))).----))) ( -28.70) >DroSim_CAF1 41113 96 + 1 AUCGAUUGGCCGAAACCCG---AU-GGGGAAAAUAGCAAUCGAUAGCU---------CAGUUCAGCUGGCCCAUUCAGCAAUUGGCCAAAAUGAAUGGUU-UGCCAA----CGA .(((.((((((((..(((.---..-)))(((...(((........)))---------...))).(((((.....)))))..))))))))......(((..-..))).----))) ( -28.50) >DroEre_CAF1 41995 100 + 1 AUCGAUUGCCCGAAACUCGCAAACUCGGGAAAAUAGCAGUCGAUAGCU---------CAGUUCAGCUGGCCCAUUCAGCAAUUGGCCAAAAUGAAUGGUU-UGCCAA----CGA (((((((((......((((......))))......)))))))))((((---------......))))((((((((((((.....)).....)))))))..-.)))..----... ( -28.90) >DroYak_CAF1 42339 106 + 1 AUCGAUUGCCCGAAACUCG---AUGGGGGAAAAUAGCAAUCGAUAGCUCAGUUAGCUCAGUUCAGCUGGCCCAUUCAGCAAUUGGCCAAAAUGAAUGGUU-UGCCAA----CGA (((((((((......(((.---...))).......)))))))))......(((.((...(((((..(((((.(((....))).)))))...)))))....-.)).))----).. ( -30.22) >DroPer_CAF1 49415 90 + 1 AUUACUGGCCUG-----UU----UGCAGGAAAAUAAUCUUCGAUGGCU---------UA-----GCCAUCCUAUUCAGCAACUGGCUA-AAUGAAUGGGUCUGGGAAAACCCAA .....(((((.(-----((----.((((((......)))).((((((.---------..-----)))))).......))))).)))))-......(((((........))))). ( -25.60) >consensus AUCGAUUGCCCGAAACCCG___AU_GGGGAAAAUAGCAAUCGAUAGCU_________CAGUUCAGCUGGCCCAUUCAGCAAUUGGCCAAAAUGAAUGGUU_UGCCAA____CGA ((((((((.......(((........))).......))))))))((((...............))))((((((((((((.....)).....)))))))....)))......... (-20.01 = -19.30 + -0.71)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:10 2006