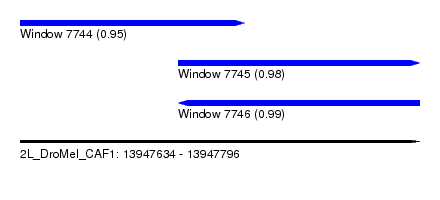
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,947,634 – 13,947,796 |
| Length | 162 |
| Max. P | 0.987459 |

| Location | 13,947,634 – 13,947,725 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.88 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13947634 91 + 22407834 UGCGCUUGUAUUGGUUAUAUUU----GUGUGUAUU--AGUGCGGCACGGAAAGUUGCAAGCGGAAGCGCU-----CACCAAUACACUGGCAUGCCACCAAUA ........(((((((.......----..(((((((--.(((.(((((.....)).....((....)))))-----))).))))))).((....))))))))) ( -26.10) >DroVir_CAF1 45331 95 + 1 UGCCUUGGUAUUGGUGAGCUUUGUGCAUUUGUGUG--CGUUGCGCUCGGAAAGUUUGCAGCGGAAGCACU-----CACUAAUACAGACGCGCCGCACUAAUA .((.((.(((((((((((((((..((((....)))--)((((((..(.....)..)))))).)))))..)-----))))))))).)).))............ ( -31.50) >DroSec_CAF1 35431 91 + 1 UGCGCUUGUAUUGGUAAUGCUU----GUGUGUAUU--AGUGCGGCACGGAAAGUUGCAAGCGGAAGCACU-----CACCAACACACUGGCAUGCCACCAAUA ........(((((((.(((((.----((((((...--..((((((.......)))))).((....))...-----.....)))))).)))))...))))))) ( -32.80) >DroEre_CAF1 36651 91 + 1 UUUGCUUGUAUUGGUAAUAUUU----GUGAGUGUU--AGUGCGGCACGGAAAGUUGCAAGCGGAAGCGCU-----CACCAAUACAUUGGCAGACCACCAAUA (((((((((((((((.......----..((((...--..((((((.......)))))).((....)))))-----))))))))))..))))))......... ( -29.40) >DroYak_CAF1 37007 91 + 1 UUCGCUUGUAUUGGUAAAAUGU----UUAAGUGUU--AGUGCCGCACGGAAAGUUGCAAGCGGAAGCGCU-----CACCAAUACAUUGGCAAAGCACCAAUA ...((((((((((((......(----((..((((.--......))))..)))......((((....))))-----.)))))))))..)))............ ( -23.20) >DroAna_CAF1 35730 94 + 1 UGCGCCUGUAUUGGUGGGUUUU----GCAGGUGUGAGAGUUUGGCUCGGAAAGUUCCAAGCGGAAGCACCACCUCUACCAAUGCAAG----AGGCACCAACA ...(((((((((((((((....----.).(((((....((((((..(.....)..))))))....)))))....))))))))))...----))))....... ( -35.30) >consensus UGCGCUUGUAUUGGUAAUAUUU____GUGUGUGUU__AGUGCGGCACGGAAAGUUGCAAGCGGAAGCACU_____CACCAAUACACUGGCAAGCCACCAAUA ...((((((((((((........................((((((.......)))))).((....)).........)))))))))..)))............ (-17.71 = -18.88 + 1.17)


| Location | 13,947,698 – 13,947,796 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 72.56 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -14.89 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13947698 98 + 22407834 CACCAAUAC------------ACUGGCAUGCCACCAAUACUCACGCAAUUGCGGCUUUUCUCUGUGUGUAUUGGUGACUUUAGCCGCCAACUUGAAAGUGGCUUAUAAAA (((((((((------------(((((....)))..........(((....)))............))))))))))).....((((((..........))))))....... ( -30.30) >DroPse_CAF1 41630 110 + 1 CACCACUACCGACACAGACAUACGAGUGUGCCACCAAUACAACGGCAAAGGCGGCUCGGCUGUGCAUGUGUGGGUAGAGCGAGCAGUCGUCUUGGAAGUGCCGCAUAAAA .....(((((.(((((..(((((((((.((((.((........))....)))))))))..))))..))))).))))).(((.(((.((......))..))))))...... ( -39.80) >DroSec_CAF1 35495 98 + 1 CACCAACAC------------ACUGGCAUGCCACCAAUACUCGCGCAAUCGCGGCUUUUCUUUCUGUGUAUUGGUGACUUUAGCCGCCGACUUGAAAGUGGCUUAUAAAA ((((((.((------------(((((....))).......(((((....)))))...........)))).)))))).....((((((..........))))))....... ( -27.10) >DroEre_CAF1 36715 98 + 1 CACCAAUAC------------AUUGGCAGACCACCAAUACUCGCGCAAUCGUGGCUUUCCUCUUUGUGUAUUGGUGACUUUAGCCGCCGACUUGAAAGUGGCUUAUAAAA (((((((((------------((.((.(((((((................)))).))))).....))))))))))).....((((((..........))))))....... ( -27.19) >DroYak_CAF1 37071 98 + 1 CACCAAUAC------------AUUGGCAAAGCACCAAUACUCGCGCAAUCGUGGAUUUUCGCUUUGUGUAUUGGUGACUUUAGCCGCCGACUUGAAAGUGGCUUAUAAAA .........------------...((((((((((((((((....((((..((((....)))).))))))))))))).)))).)))(((.(((....))))))........ ( -29.90) >DroPer_CAF1 43553 110 + 1 CACCACUACCGACACAGACAUACGAGUGUGCCACCAAUACAACGGCAAAGGCGGCUCGGCUGUGCAUGUGUGGGUAGAGCGAGCAGUCGUCUUGGAAGUGCCGCAUAAAA .....(((((.(((((..(((((((((.((((.((........))....)))))))))..))))..))))).))))).(((.(((.((......))..))))))...... ( -39.80) >consensus CACCAAUAC____________ACUGGCAUGCCACCAAUACUCGCGCAAUCGCGGCUUUUCUCUGUGUGUAUUGGUGACUUUAGCCGCCGACUUGAAAGUGGCUUAUAAAA ........................(((.((.(((((((((...(((....)))((........))..)))))))))...)).)))((((.((....))))))........ (-14.89 = -15.33 + 0.45)



| Location | 13,947,698 – 13,947,796 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.56 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -18.16 |
| Energy contribution | -19.63 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13947698 98 - 22407834 UUUUAUAAGCCACUUUCAAGUUGGCGGCUAAAGUCACCAAUACACACAGAGAAAAGCCGCAAUUGCGUGAGUAUUGGUGGCAUGCCAGU------------GUAUUGGUG .......((((.((........)).)))).....(((((((((((...(.(....(((((...(((....)))...)))))...)).))------------))))))))) ( -34.60) >DroPse_CAF1 41630 110 - 1 UUUUAUGCGGCACUUCCAAGACGACUGCUCGCUCUACCCACACAUGCACAGCCGAGCCGCCUUUGCCGUUGUAUUGGUGGCACACUCGUAUGUCUGUGUCGGUAGUGGUG ......((((((..((......)).))).))).(((((.(((((.(.(((..(((((((((..(((....)))..))))))....)))..))))))))).)))))..... ( -37.00) >DroSec_CAF1 35495 98 - 1 UUUUAUAAGCCACUUUCAAGUCGGCGGCUAAAGUCACCAAUACACAGAAAGAAAAGCCGCGAUUGCGCGAGUAUUGGUGGCAUGCCAGU------------GUGUUGGUG ........((((((....))).)))(((....)))((((((((((..........(((((.(.(((....))).).)))))......))------------)))))))). ( -33.89) >DroEre_CAF1 36715 98 - 1 UUUUAUAAGCCACUUUCAAGUCGGCGGCUAAAGUCACCAAUACACAAAGAGGAAAGCCACGAUUGCGCGAGUAUUGGUGGUCUGCCAAU------------GUAUUGGUG ........((((((....))).)))(((....)))(((((((((......((...(((((.(.(((....))).).)))))...))..)------------)))))))). ( -32.90) >DroYak_CAF1 37071 98 - 1 UUUUAUAAGCCACUUUCAAGUCGGCGGCUAAAGUCACCAAUACACAAAGCGAAAAUCCACGAUUGCGCGAGUAUUGGUGCUUUGCCAAU------------GUAUUGGUG ........((((((....))).)))(((.((((.(((((((((.....(((..((((...)))).)))..))))))))))))))))...------------......... ( -31.00) >DroPer_CAF1 43553 110 - 1 UUUUAUGCGGCACUUCCAAGACGACUGCUCGCUCUACCCACACAUGCACAGCCGAGCCGCCUUUGCCGUUGUAUUGGUGGCACACUCGUAUGUCUGUGUCGGUAGUGGUG ......((((((..((......)).))).))).(((((.(((((.(.(((..(((((((((..(((....)))..))))))....)))..))))))))).)))))..... ( -37.00) >consensus UUUUAUAAGCCACUUUCAAGUCGGCGGCUAAAGUCACCAAUACACACAGAGAAAAGCCGCGAUUGCGCGAGUAUUGGUGGCAUGCCAGU____________GUAUUGGUG ........((((((....))).))).........(((((((((.(.....)....(((((.(.(((....))).).)))))....................))))))))) (-18.16 = -19.63 + 1.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:07 2006