| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,929,499 – 13,929,604 |
| Length | 105 |
| Max. P | 0.877349 |

| Location | 13,929,499 – 13,929,604 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.66 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.19 |
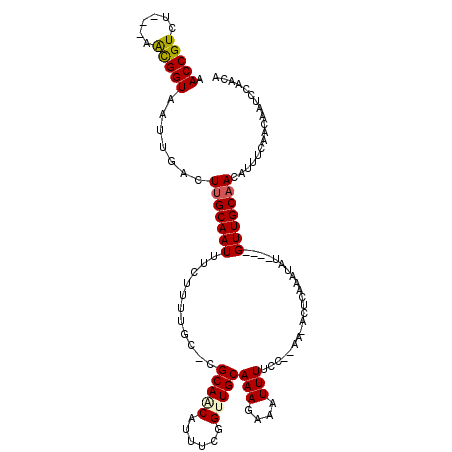
| Structure conservation index | 0.67 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
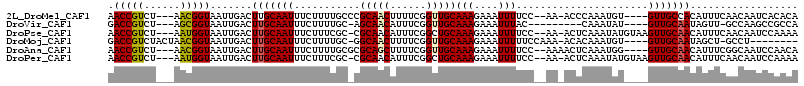
>2L_DroMel_CAF1 13929499 105 - 22407834 AACCGUCU---AACGGUAAUUGACUUGCAAUUUCUUUUGCCCGCAACUUUUCGGUUGCAAAGAAAUUUUCC--AA-ACCCAAAUGU----GUUGCCACAUUUCAACAAUCACACA .((((...---..))))...(((.(((.(((((((((.....((((((....)))))))))))))))....--..-....((((((----(....)))))))...)))))).... ( -21.30) >DroVir_CAF1 19604 97 - 1 GACCGUCU---AGCGGUAAUUGACUUGCAAUUUCUUUUGC-AGCAACAUUUCGGUUGCAAAGAAAUUUAC---------CAAAUAU----GUUGCAAUAGUU-GCCAAGCCGCCA ........---.(((((....(((((((((......))))-)(((((((...(((...(((....)))))---------)....))----)))))...))))-.....))))).. ( -25.40) >DroPse_CAF1 16669 108 - 1 AACCGUCU---AAUGGUAAUUGACUUGCAAUUUCUUUCGC-CGCAACAUUUCGGCUGCAAAGAAAUUUUCC--AA-ACUCAAAUAUGUAAGUUGCAACAUUUCAACAAUCCAAAA .(((((..---.)))))....(((((((((((((((((((-((........)))).).)))))))).....--..-.........))))))))...................... ( -19.09) >DroMoj_CAF1 18495 100 - 1 GACCGUCUACUAACGGUAAUUGACUUGCAAUUUCUUUUGC-GGCAACUUUUCGGUUGCAAAGAAAUUUUUCCAAA-ACACAAAUGU----GUUGCAAUAGCU-GCCU-------- .(((((......))))).......(((((((((((((.((-(((.........)))))))))))))).......(-((((....))----))))))).....-....-------- ( -24.40) >DroAna_CAF1 18257 106 - 1 AACCGUCU---AACGGUAAUUGACUUGCAAUUUCUUUUGCGCGCAGCUUUUCGGUUGCAAAGAAAUUUUCC--AAAACUCAAAUGG----GUUGCAACAUUUCGGCAAUCCAACA .((((...---..))))..((((...((((......))))..((((((....)))))).............--.....)))).(((----(((((.........))))))))... ( -27.50) >DroPer_CAF1 18899 108 - 1 AACCGUCU---AAUGGUAAUUGACUUGCAAUUUCUUUCGC-CGCAACAUUUCGGCUGCAAAGAAAUUUUCC--AA-ACUCAAAUAUGUAAGUUGCAACAUUUCAACAAUCCAAAA .(((((..---.)))))....(((((((((((((((((((-((........)))).).)))))))).....--..-.........))))))))...................... ( -19.09) >consensus AACCGUCU___AACGGUAAUUGACUUGCAAUUUCUUUUGC_CGCAACAUUUCGGUUGCAAAGAAAUUUUCC__AA_ACUCAAAUAU____GUUGCAACAUUUCAACAAUCCAACA .(((((......))))).......(((((((...........(((((......)))))(((....)))......................))))))).................. (-15.36 = -15.42 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:30:03 2006