| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,445,504 – 1,445,643 |
| Length | 139 |
| Max. P | 0.856016 |

| Location | 1,445,504 – 1,445,608 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.03 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -11.23 |
| Energy contribution | -12.73 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
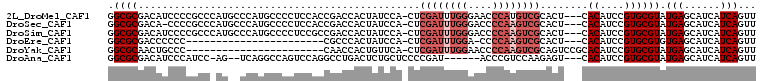
>2L_DroMel_CAF1 1445504 104 - 22407834 GGCGCGACAUCCCCGCCCAUGCCCAUGCCCCUCCACCGACCACUAUCCA-CUCGAUUUGGGAACCCAUGUCGCACU---CACAUCCGUGCGUAUGAGCAUCAUCAGUU ((.(((.......)))))((((.(((((.....................-...(((.(((....))).)))((((.---.......))))))))).))))........ ( -27.60) >DroSec_CAF1 63519 103 - 1 GGCGCGACA-CCCCGCCCAUGCCCAUGCCCCUCCACCGACCACUAUCCA-CUCGAUUUGGGACCCCAAGUCGCACU---CACAUCCGUGCGUAUGAGCAUCAUCAGUU ((.(((...-...)))))((((.(((((.....................-...(((((((....)))))))((((.---.......))))))))).))))........ ( -31.30) >DroSim_CAF1 66037 104 - 1 GGCGCGACAUCCCCGCCCAUGCCCAUGCCCCUCCGCCGACCACUAUCCA-CUCGAUUUGGGACCCCAAGUCGCACU---CACAUCCGUGCGUAUGAGCAUCAUCAGUU ((.(((.......)))))((((.(((((.....................-...(((((((....)))))))((((.---.......))))))))).))))........ ( -30.80) >DroEre_CAF1 66019 80 - 1 GGCGCGACCCCCC-----------------------CGCCCACUAUCCA-CUCGAUUUGGA-CCCCAAGUCGCACU---CACAUCCGUGCGUGUGAGCAUCAUCAGUU ((.(((.......-----------------------)))))........-((((((((((.-..)))))))((((.---(((....))).)))))))........... ( -22.90) >DroYak_CAF1 64903 84 - 1 GGCGCAACUGCCC-----------------------CAACCACUGUUCA-CUCGAUUUGGAACCCCAAGUCGCAGUCCGCACAUCCGUGCGUAUGAGCAUCAUCAGUU ((.((....)).)-----------------------)......((((((-((((((((((....)))))))).))).(((((....)))))...)))))......... ( -27.70) >DroAna_CAF1 63800 96 - 1 GGCGCGACAUCCCAUCC-AG--UCAGGCCAGUCCAGGCCUGACUCUGCUCCCCGAU------ACCCGUCCAAGAGU---CACAUCCGUGCGUAUGAGCAUCAUCAGUU .(((((...........-((--(((((((......)))))))))..((((...(((------....)))...))))---......)))))...(((......)))... ( -29.20) >consensus GGCGCGACAUCCCCGCC_AU__CCAUGCCCCUCCACCGACCACUAUCCA_CUCGAUUUGGGACCCCAAGUCGCACU___CACAUCCGUGCGUAUGAGCAUCAUCAGUU .((((...............................................((((((((....))))))))........((....)))))).(((......)))... (-11.23 = -12.73 + 1.50)
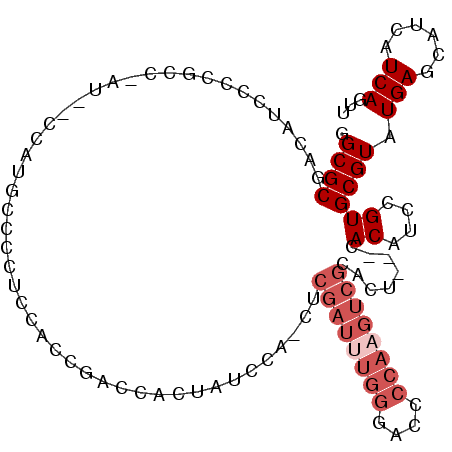
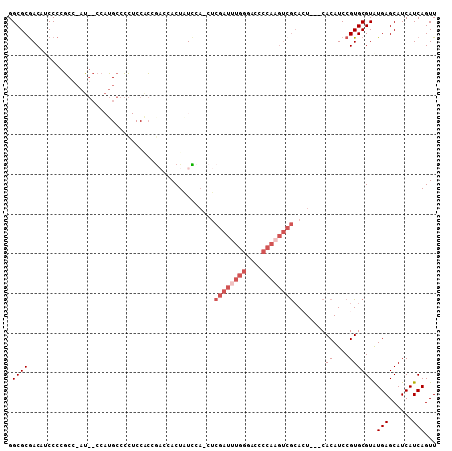
| Location | 1,445,533 – 1,445,643 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.02 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -16.18 |
| Energy contribution | -17.68 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1445533 110 - 22407834 CAGGUCUAAUCGCCUUGGCGACACCUAUUUUCGUAGGCGCGACAUCCCCGCCCAUGCCCAUGCCCCUCCACCGACCACUAUCCA-CUCGAUUUGGGAACCCAUGUCGCACU ..((((...((((....))))...........(((((.(((.......))))).)))...............))))........-..((((.(((....))).)))).... ( -27.30) >DroSec_CAF1 63548 109 - 1 CAGGCCUAAUCGCCUUGGCGACACCUAUUUUCGUAGGCGCGACA-CCCCGCCCAUGCCCAUGCCCCUCCACCGACCACUAUCCA-CUCGAUUUGGGACCCCAAGUCGCACU ..(((....((((....))))...........(((((.(((...-...))))).)))....)))....................-..((((((((....)))))))).... ( -28.20) >DroSim_CAF1 66066 110 - 1 CAGGUCUAAUCGCCUUGGCGACACCUAUUUUCGUAGGCGCGACAUCCCCGCCCAUGCCCAUGCCCCUCCGCCGACCACUAUCCA-CUCGAUUUGGGACCCCAAGUCGCACU ..((((...((((....))))..............((((((.......))).(((....))).......)))))))........-..((((((((....)))))))).... ( -31.40) >DroEre_CAF1 66048 86 - 1 CAGGUCUAAUCGCCUUGGCGACACCUAUUUUCGUAGGCGCGACCCCCC-----------------------CGCCCACUAUCCA-CUCGAUUUGGA-CCCCAAGUCGCACU .((((....((((....)))).))))......((.((((.(......)-----------------------)))).))......-..((((((((.-..)))))))).... ( -24.10) >DroYak_CAF1 64935 87 - 1 CAGGUCUAAUCGCCUUGGCGACACCUAUUUUCGUAGGCGCAACUGCCC-----------------------CAACCACUGUUCA-CUCGAUUUGGAACCCCAAGUCGCAGU .((((....((((....)))).))))......((.((.((....)).)-----------------------).))........(-((((((((((....)))))))).))) ( -25.20) >DroAna_CAF1 63829 102 - 1 CAGGCCUAAUCGCCUUGGCGACACCUAUUUUCGUAGGCGCGACAUCCCAUCC-AG--UCAGGCCAGUCCAGGCCUGACUCUGCUCCCCGAU------ACCCGUCCAAGAGU ((((((((.((((....))))............)))))..............-((--(((((((......))))))))))))(((...(((------....)))...))). ( -31.32) >consensus CAGGUCUAAUCGCCUUGGCGACACCUAUUUUCGUAGGCGCGACAUCCCCGCC_AU__CCAUGCCCCUCCACCGACCACUAUCCA_CUCGAUUUGGGACCCCAAGUCGCACU ..((......(((((..((((.........)))))))))......))........................................((((((((....)))))))).... (-16.18 = -17.68 + 1.50)

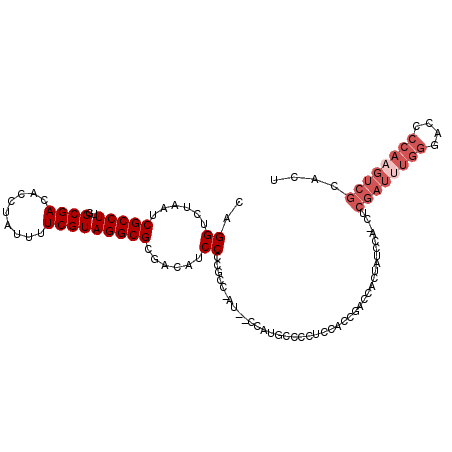
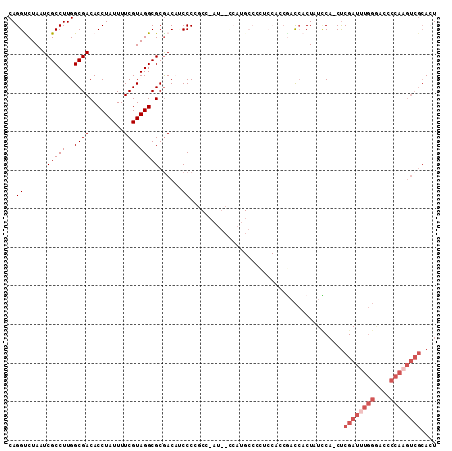
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:22 2006