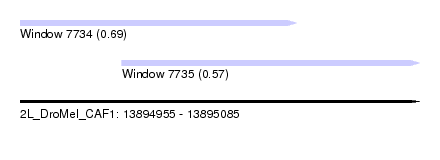
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,894,955 – 13,895,085 |
| Length | 130 |
| Max. P | 0.687264 |

| Location | 13,894,955 – 13,895,045 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -13.78 |
| Consensus MFE | -10.37 |
| Energy contribution | -10.65 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13894955 90 + 22407834 CUUUCCCUUGCCAAUUGCCAACCCUUUGCCAUUUUAAAUUCAUUCUAUGAUUUUUCGUCUUUUUCCCUUACAGUUCGCUGGCUGGGUGGA .................(((.(((...(((................((((....)))).............((....))))).)))))). ( -13.90) >DroSec_CAF1 38686 90 + 1 CUUUCCCUUGCCAAUUGCCAACCCUUUGCCAUUUUAUACUCAUUAUAUGAUUUUUCGUCUUUUUCCCUUACAGUUCGCUGGCUGGGUGGA .................(((.(((...(((................((((....)))).............((....))))).)))))). ( -13.90) >DroSim_CAF1 40939 90 + 1 CUUUCCCUUGCCAAUUGCCAACCCUUUGCCAUUUUAUACUCAUUAUAUGAUUUUUCGUCUUUUUCCCUUGCAGUUCGCUGGCUGGGUGGA .................(((.(((...((((...............((((....))))...........((.....)))))).)))))). ( -15.80) >DroEre_CAF1 38803 90 + 1 CUUUCCCUUCCCAACUGCCAACCCUUUGCCAUUCCUAACUCAUUAUAUGAUUUUUCGUCCUUUUUCCUUACAGUUCGCUGGCUGGGUGGA .................(((.(((...(((................((((....)))).............((....))))).)))))). ( -13.90) >DroYak_CAF1 40872 73 + 1 ----------------GCCAACCCUUUGCCGUUCUUAACUCAUUAUAUGAUUUUUCGUC-UUUUUCCUUACAGUUCGCUGGCUGGGUGGA ----------------.(((.(((...(((................((((....)))).-...........((....))))).)))))). ( -13.90) >DroPer_CAF1 49792 73 + 1 A--UCCCUUG-----UGC---CCCUUUCUCAUGCAUACCU-------UUCCCUUGCACACAUUCUCCUUACAGUUCGCUGGCUGGGUGGA .--(((....-----.((---((........((((.....-------......)))).............(((....)))...))))))) ( -11.30) >consensus CUUUCCCUUGCCAAUUGCCAACCCUUUGCCAUUCUAAACUCAUUAUAUGAUUUUUCGUCUUUUUCCCUUACAGUUCGCUGGCUGGGUGGA .................(((.(((...(((................((((....)))).............((....))))).)))))). (-10.37 = -10.65 + 0.28)

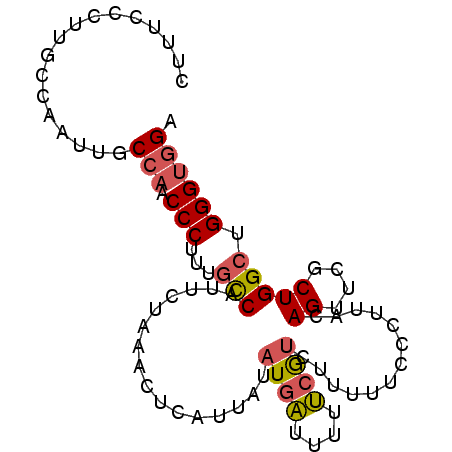
| Location | 13,894,988 – 13,895,085 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -19.76 |
| Energy contribution | -18.85 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13894988 97 + 22407834 UUAAAUUCAUUCUA-------UGAUUUUUCGUC----UUUUUCCCUUACAGUUCGCUGGCUGGGUGGAUGCGGUAAUCUUUGUGUUCAGCCUGGAGAACGAGGGCAGC ..............-------.........(((----((((((((...(((....)))...((.(((((((((......))))))))).)).)).))).))))))... ( -23.70) >DroGri_CAF1 46607 100 + 1 UUUAACUAAUUCUCUUUCGCUCUCGUUGCGCUCU--------UUCUUUCAGUUCGCUGGCUGGGUGGAUGCGGUAAUCUUUGUGUUUAGCCUGGAGAACGAGGGCAGC ..................((((((((((((..((--------.......))..)))...(..(((((((((((......))))))))).))..)..)))))))))... ( -29.20) >DroEre_CAF1 38836 97 + 1 CCUAACUCAUUAUA-------UGAUUUUUCGUC----CUUUUUCCUUACAGUUCGCUGGCUGGGUGGAUGCGGUAAUCUUCGUGUUCAGUCUGGAGAACGAGGGCAGC ..............-------.........(((----((((((((...(((....)))...((.(((((((((......))))))))).)).)))))..))))))... ( -26.90) >DroWil_CAF1 54722 100 + 1 U--CACUCAUUUCGUUUUU--CUCUCUUUCUCA----CUUUAUCUUUACAGUUCGCUGGCUGGGUGGAUGCGGUAAUAUUUGUAUUUAGCCUGGAAAACGAGGGCAGU .--.(((..((((((((((--(...........----...........(((....)))...((.(((((((((......))))))))).)).)))))))))))..))) ( -24.90) >DroYak_CAF1 40889 96 + 1 CUUAACUCAUUAUA-------UGAUUUUUCGUC-----UUUUUCCUUACAGUUCGCUGGCUGGGUGGAUGCGGUAAUCUUCGUGUUCAGCCUGGAAAACGAGGGCAGC .....(((.....(-------(((....)))).-----.((((((...(((....)))...((.(((((((((......))))))))).)).)))))).)))...... ( -25.60) >DroAna_CAF1 36081 94 + 1 UU---CUCA---UA-------CUAUUUUGCAUC-CUCUCGUAUCCUUACAGUUCGCUGGCUGGGUGGAUGCCGUAAUCUUCGUGUUCAGCCUGGAGAACGAGGGCAGU ..---....---..-------.....((((..(-(((..(((....))).((((.(((((((..(((((......))))....)..))))).)).)))))))))))). ( -27.50) >consensus UUUAACUCAUUAUA_______CGAUUUUUCGUC_____UUUUUCCUUACAGUUCGCUGGCUGGGUGGAUGCGGUAAUCUUCGUGUUCAGCCUGGAGAACGAGGGCAGC ..........................................(((((.(((....))).(..(((((((((((......))))))))).))..).....))))).... (-19.76 = -18.85 + -0.91)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:56 2006