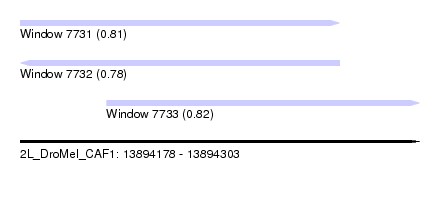
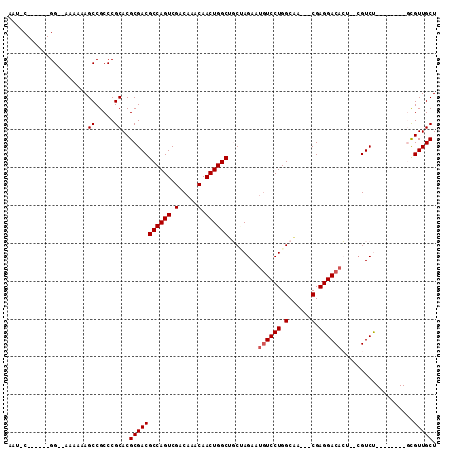
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,894,178 – 13,894,303 |
| Length | 125 |
| Max. P | 0.816973 |

| Location | 13,894,178 – 13,894,278 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -42.19 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.17 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13894178 100 + 22407834 AGCAACGC--------AGACG--AGUGUCCUCG---UUGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUUUCCCAUUGCCGAAAG .(((.(((--------.((((--((((((((..---.....))))))))).....((((((((.....)))))))))))))))))...((((.............)))).... ( -43.62) >DroPse_CAF1 47018 96 + 1 AGCAACGA--------GGACGCCGUUGUCCUCGUCGUUGCCAGGACAUUCUCGAAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CC------G-AUU ....((((--------(((((....))))))))).((((((..(.(((...(((.((((((((.....)))))))).))).)))).))))))......--..------.-... ( -39.80) >DroEre_CAF1 38051 98 + 1 AGCAACGC--------AGACG--AGUGUCCUCG---UCGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CCAUCGCCGAACU .(((.(((--------.((((--((((((((..---.....))))))))).....((((((((.....)))))))))))))))))(..((((......--.....))))..). ( -45.30) >DroYak_CAF1 40112 98 + 1 AGCAACGC--------AGACG--AGUGUCCUCG---UCGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CCAUUGCCGAACU .(((.(((--------.((((--((((((((..---.....))))))))).....((((((((.....)))))))))))))))))(..((((......--.....))))..). ( -45.90) >DroAna_CAF1 35316 100 + 1 AGCAACACAGGACAGCAGAC---UGUGUCCUCG---UUGCCAGGAUGUUCUGGCUGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUUCUCC------G-GGU .(((((..((((((......---..)))))).)---))))........(((((..((((((((.....))))))))(((((......)))))........))------)-)). ( -38.70) >DroPer_CAF1 48897 96 + 1 AGCAACGA--------GGACGCCGUUGUCCUCGUCGUUGCCAGGACAUUCUCGAAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CC------G-AUU ....((((--------(((((....))))))))).((((((..(.(((...(((.((((((((.....)))))))).))).)))).))))))......--..------.-... ( -39.80) >consensus AGCAACGC________AGACG__AGUGUCCUCG___UUGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU__CC______G_ACU .........................((((((.(......).))))))........((((((((.....))))))))(((((......)))))..................... (-27.44 = -27.17 + -0.28)

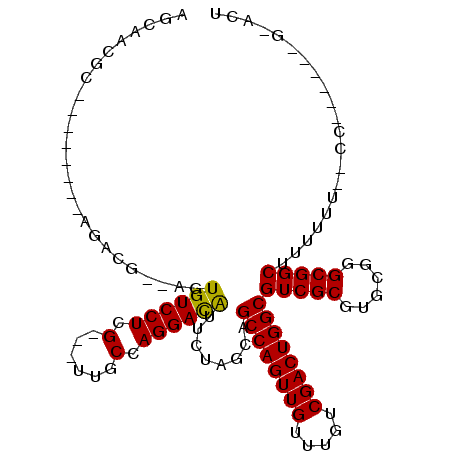
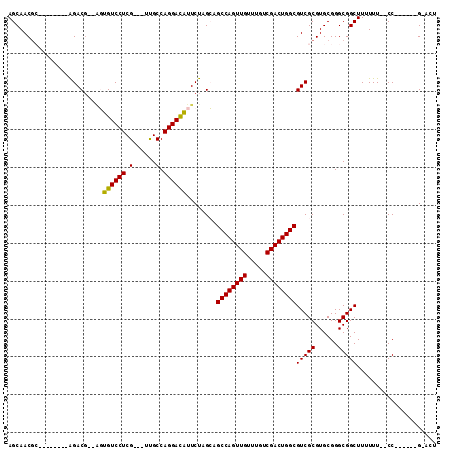
| Location | 13,894,178 – 13,894,278 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13894178 100 - 22407834 CUUUCGGCAAUGGGAAAAAAAGCCGCCCGCACGCGACGCCAGUCGACAAACAACUGGCUGCUAGAAUGUCCUGGCAA---CGAGGACACU--CGUCU--------GCGUUGCU ....((((.............))))...(((((((((((((((.(.....).)))))).....((.(((((((....---).)))))).)--)))).--------))).))). ( -37.42) >DroPse_CAF1 47018 96 - 1 AAU-C------GG--AAAAAAGCCGCCCGCACGCGACGCCAGUCGACAAACAACUGGCUUCGAGAAUGUCCUGGCAACGACGAGGACAACGGCGUCC--------UCGUUGCU ...-.------..--......((((...(((..(((.((((((.(.....).)))))).)))....)))..))))..((((((((((......))))--------)))))).. ( -36.20) >DroEre_CAF1 38051 98 - 1 AGUUCGGCGAUGG--AAAAAAGCCGCCCGCACGCGACGCCAGUCGACAAACAACUGGCUGCUAGAAUGUCCUGGCGA---CGAGGACACU--CGUCU--------GCGUUGCU .(..((((.....--......))))..)(((((((..((((((.(.....).)))))).(((((......)))))((---((((....))--)))))--------))).))). ( -39.80) >DroYak_CAF1 40112 98 - 1 AGUUCGGCAAUGG--AAAAAAGCCGCCCGCACGCGACGCCAGUCGACAAACAACUGGCUGCUAGAAUGUCCUGGCGA---CGAGGACACU--CGUCU--------GCGUUGCU .(..((((.....--......))))..)(((((((..((((((.(.....).)))))).(((((......)))))((---((((....))--)))))--------))).))). ( -39.10) >DroAna_CAF1 35316 100 - 1 ACC-C------GGAGAAAAAAGCCGCCCGCACGCGACGCCAGUCGACAAACAACUGGCAGCCAGAACAUCCUGGCAA---CGAGGACACA---GUCUGCUGUCCUGUGUUGCU ...-(------((.(........)..)))...(((((((((((.(.....).)))))).(((((......))))).(---(.((((((..---......))))))))))))). ( -34.30) >DroPer_CAF1 48897 96 - 1 AAU-C------GG--AAAAAAGCCGCCCGCACGCGACGCCAGUCGACAAACAACUGGCUUCGAGAAUGUCCUGGCAACGACGAGGACAACGGCGUCC--------UCGUUGCU ...-.------..--......((((...(((..(((.((((((.(.....).)))))).)))....)))..))))..((((((((((......))))--------)))))).. ( -36.20) >consensus AAU_C______GG__AAAAAAGCCGCCCGCACGCGACGCCAGUCGACAAACAACUGGCUGCUAGAAUGUCCUGGCAA___CGAGGACACU__CGUCU________GCGUUGCU .....................((.....))..(((((((((((.(.....).))))))........((((((.(......).))))))...................))))). (-24.58 = -24.92 + 0.33)

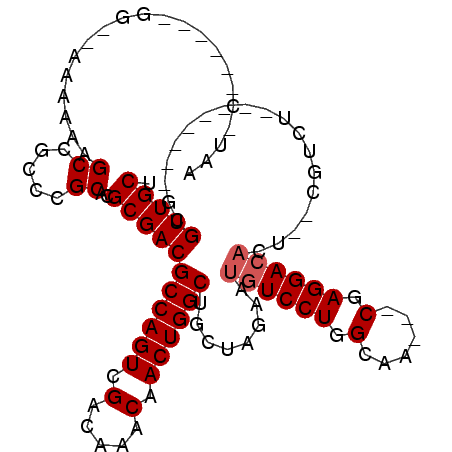
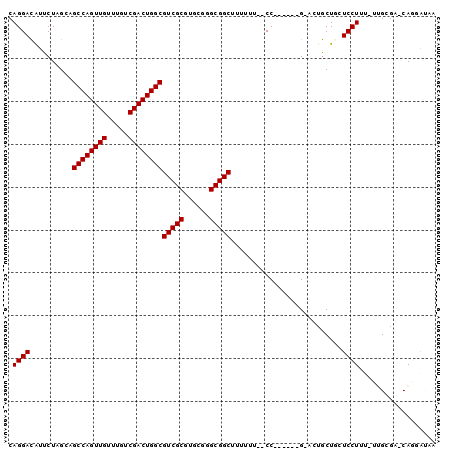
| Location | 13,894,205 – 13,894,303 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13894205 98 + 22407834 CAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUUUCCCAUUGCCGAAAGGCUGCUCCUUU-UUGUGU-CAGGAUAA (..(((((...((((((((((((.....))))))))(((((......))))).............(((....))))))).....-..))))-)..).... ( -36.30) >DroPse_CAF1 47050 84 + 1 CAGGACAUUCUCGAAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CC------G-AUUGCUCCUCCU-----GCGA-CAUCG-AC ..........((((.((((((((.....))))))))((((((.(.(((.(((..((..--..------)-)..))).)))))-----))))-).)))-). ( -32.20) >DroEre_CAF1 38078 96 + 1 CAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CCAUCGCCGAACUGCUGCUCCUUU-UUGGGG-CAGGAUAA ..(((..........((((((((.....))))))))(((((......))))).....)--))............(((((((...-..))))-)))..... ( -36.70) >DroYak_CAF1 40139 97 + 1 CAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CCAUUGCCGAACUGCUGCUCCUUU-UUGGGGACAGGAUAA ......(((((....((((((((.....))))))))((((((.((((.((((......--.....))))..))))))).(((..-..))))))))))).. ( -36.00) >DroAna_CAF1 35350 92 + 1 CAGGAUGUUCUGGCUGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUUCUCC------G-GGUACUAGUCCUCUGCUGUGA-CAGGAUAA .....((((((....((((((((.....))))))))((((((.(((((.((((....((...------.-))....)))).))))))))))-))))))). ( -36.20) >DroPer_CAF1 48929 84 + 1 CAGGACAUUCUCGAAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CC------G-AUUGCUCCUCCU-----GCGA-CAUCG-AC ..........((((.((((((((.....))))))))((((((.(.(((.(((..((..--..------)-)..))).)))))-----))))-).)))-). ( -32.20) >consensus CAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU__CC______G_ACUGCUGCUCCUUU_UUGCGA_CAGGAUAA .((((..........((((((((.....))))))))(((((......)))))..........................)))).................. (-24.27 = -24.27 + 0.00)

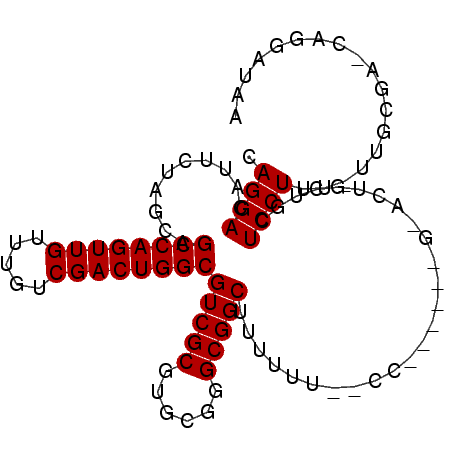
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:55 2006