| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,867,467 – 13,867,560 |
| Length | 93 |
| Max. P | 0.708562 |

| Location | 13,867,467 – 13,867,560 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.45 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13867467 93 + 22407834 -----------AAG-----AGGAGCAGCUC----UGCAUCGACUUCAAUGUAGAGCAAAACUCAUUUAGUAGCUGCUGC---UGUUGCUGAUGUUUACUAUGUUAUCCUUUUU--AUG-- -----------(((-----(((((((((((----(((((.(....).))))))))).((((....((((((((......---.)))))))).))))....)))..))))))).--...-- ( -30.00) >DroSec_CAF1 11535 96 + 1 -----------AAG-----AGGAGCAGCAC----UGCAUCGACUUCAAUGUGGAGCAAAACUCAUUUAGUAGCUGCUGCUCCUGCUGCUGAUGUUUACUAUGUUAUCCUUUUU--AUG-- -----------.((-----(((((((((((----(((....((......)).(((.....))).....)))).)))))))))).))..((((((.....))))))........--...-- ( -26.80) >DroSim_CAF1 13046 96 + 1 -----------AAG-----AGGAGCAGUUC----UGCAUCGACUUCAAUGUAGAGCAAAACUCAUUUAGUAGCUGCUGCUCCUGCUGCUGAUGUUUACUAUGUUAUCCUUUUU--AUG-- -----------(((-----(((((((((((----(((((.(....).))))))))).((((....((((((((..........)))))))).))))....)))..))))))).--...-- ( -29.40) >DroEre_CAF1 11842 98 + 1 GAUGCAGGAUAAAG-----AGGAGCAGCUC----UGCAUCGACUUCAAUGUAGAGCAAAACUCAUUUAGUAGCUGCC---------GCUGAUGUUUACUAUGUUAUCCUUUUU--AUG-- .....(((((((..-----(((((((((((----(((((.(....).))))))))).............((((....---------)))).))))).))...)))))))....--...-- ( -28.60) >DroYak_CAF1 13210 105 + 1 GAUGCAGGAUAAAGAGAAGAGGAGCAGCUC----UGCAUCGGCUUCAAUGUAGAGCAAAACUCAUUUAGUAGCUGCU---------GCUGAUGUUUACUAUGUUAUUCUUUUU--AUGUG ......(.((((((((((....((((((((----(((((........))))))))).((((....(((((((...))---------))))).))))....)))).))))))))--)).). ( -30.50) >DroAna_CAF1 10584 108 + 1 GAGG-------GCGCGGAGAGGAGUGCCUCCUGCAGCAGCAGCUUCAAUGUAGAGCAAAACUCAUUUAGUAGCUCCUGU---UGAUGUUGAUAUUUACUGUGUUAUUCCUUUUAUAUG-- ((((-------((((((.((((....)))))))).))..((((.(((((...((((...(((.....))).))))..))---))).)))).................)))).......-- ( -29.10) >consensus ___________AAG_____AGGAGCAGCUC____UGCAUCGACUUCAAUGUAGAGCAAAACUCAUUUAGUAGCUGCUGC___UG_UGCUGAUGUUUACUAUGUUAUCCUUUUU__AUG__ ....................((((((((......(((((.(....).)))))(((.....)))........)))))............((((((.....)))))))))............ (-12.50 = -12.45 + -0.05)

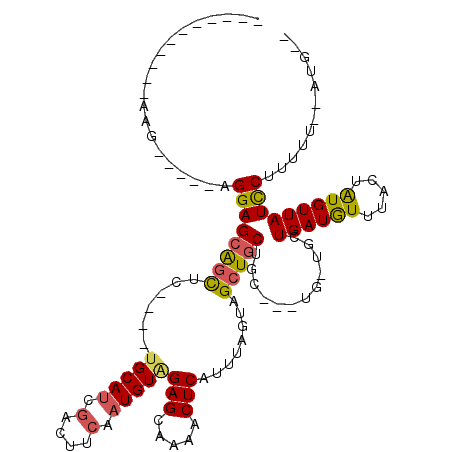
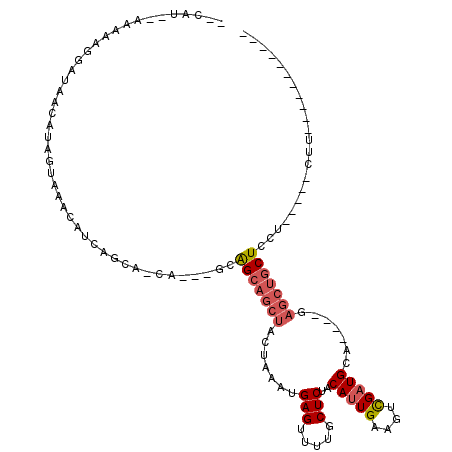
| Location | 13,867,467 – 13,867,560 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -11.48 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13867467 93 - 22407834 --CAU--AAAAAGGAUAACAUAGUAAACAUCAGCAACA---GCAGCAGCUACUAAAUGAGUUUUGCUCUACAUUGAAGUCGAUGCA----GAGCUGCUCCU-----CUU----------- --...--....((((.................((....---)).((((((.((....(((.....)))..(((((....))))).)----)))))))))))-----...----------- ( -21.20) >DroSec_CAF1 11535 96 - 1 --CAU--AAAAAGGAUAACAUAGUAAACAUCAGCAGCAGGAGCAGCAGCUACUAAAUGAGUUUUGCUCCACAUUGAAGUCGAUGCA----GUGCUGCUCCU-----CUU----------- --...--....((((.......(....)....(((((((((((((.((((........))))))))))).(((((....)))))..----.))))))))))-----...----------- ( -28.70) >DroSim_CAF1 13046 96 - 1 --CAU--AAAAAGGAUAACAUAGUAAACAUCAGCAGCAGGAGCAGCAGCUACUAAAUGAGUUUUGCUCUACAUUGAAGUCGAUGCA----GAACUGCUCCU-----CUU----------- --...--...........................((.((((((((..((.(((.....)))...))(((.(((((....))))).)----)).))))))))-----)).----------- ( -22.70) >DroEre_CAF1 11842 98 - 1 --CAU--AAAAAGGAUAACAUAGUAAACAUCAGC---------GGCAGCUACUAAAUGAGUUUUGCUCUACAUUGAAGUCGAUGCA----GAGCUGCUCCU-----CUUUAUCCUGCAUC --...--....(((((((..((((.......(((---------....)))))))...((((...(((((.(((((....))))).)----)))).))))..-----..)))))))..... ( -27.01) >DroYak_CAF1 13210 105 - 1 CACAU--AAAAAGAAUAACAUAGUAAACAUCAGC---------AGCAGCUACUAAAUGAGUUUUGCUCUACAUUGAAGCCGAUGCA----GAGCUGCUCCUCUUCUCUUUAUCCUGCAUC ...((--(((.((((.....((((.......(((---------....)))))))...((((...(((((.(((((....))))).)----)))).))))...)))).)))))........ ( -21.91) >DroAna_CAF1 10584 108 - 1 --CAUAUAAAAGGAAUAACACAGUAAAUAUCAACAUCA---ACAGGAGCUACUAAAUGAGUUUUGCUCUACAUUGAAGCUGCUGCUGCAGGAGGCACUCCUCUCCGCGC-------CCUC --........(((......................(((---(..(((((.(((.....)))...)))))...)))).(((((....)))(((((......)))))))..-------))). ( -22.20) >consensus __CAU__AAAAAGGAUAACAUAGUAAACAUCAGCA_CA___GCAGCAGCUACUAAAUGAGUUUUGCUCUACAUUGAAGUCGAUGCA____GAGCUGCUCCU_____CUU___________ ...........................................(((((((.......(((.....)))..(((((....))))).......)))))))...................... (-11.48 = -12.20 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:48 2006