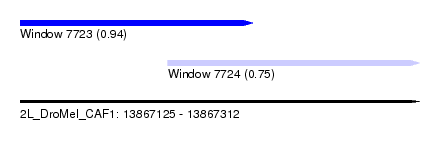
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,867,125 – 13,867,312 |
| Length | 187 |
| Max. P | 0.944222 |

| Location | 13,867,125 – 13,867,234 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13867125 109 + 22407834 CCUUAUGCCCAAUUAUAUCAAAUCGAUCGUCACAGAUCAUCAAAUCGAUCUAAGCCAAUUACACAGGCAGCUGAUUGAGUCUGGGCGAUUGUGACUGUCUCGUAAAUCU ......(((((.............((((......))))......((((((...(((.........)))....))))))...)))))((((...((......)).)))). ( -25.50) >DroSec_CAF1 11193 109 + 1 CCUUAUGCCCAAUUAUAUCAAAUCGAUCGUCACAGAUCAUCAAAUCGAUCUAAGCCAAUUACACAGGAAGCUGAUUGAGUUUGGGCGAUUGCGACUGUCUCGUAAAUCU ......((((((............((((......))))......((((((....((.........)).....))))))..))))))((((((((.....)))).)))). ( -26.60) >DroSim_CAF1 12704 109 + 1 CCUUAUGCCCAAUUAUAUCAAAUCGAUCGUCACAGAUCAUCAAAUCGAUCUAAGCCAAUUACACAGGCAGCUGAUUGAGUCUGGGCGAUUGUGACUGUCUCGUAAAUCU ......(((((.............((((......))))......((((((...(((.........)))....))))))...)))))((((...((......)).)))). ( -25.50) >DroEre_CAF1 11493 109 + 1 CCCUUUGCCCACUUACAUCAGAUCGAUCGUCACAGAUCAUCAAAUCGAUCUAAGCCAAUUACACAGGCAGCUGAUUGAGUCUGAGCGAUUGUGACUGUCUCGUAAAUCA ...(((((...((......))...(((.(((((((.((.(((..((((((...(((.........)))....))))))...)))..))))))))).)))..)))))... ( -25.10) >DroYak_CAF1 12860 109 + 1 CCCUUUGCCCAACUCUAUCAAAUCGAUCGUCACAGAUCAUCAAAUCGAUCUAAGCCAAUUACACAGGCAGCUGAUUGAGUUUGGGCGAUUGUGACUGUCUCGUAAAUCU ......(((((((((.((((....((((......))))...............(((.........)))...)))).))).))))))((((...((......)).)))). ( -28.50) >consensus CCUUAUGCCCAAUUAUAUCAAAUCGAUCGUCACAGAUCAUCAAAUCGAUCUAAGCCAAUUACACAGGCAGCUGAUUGAGUCUGGGCGAUUGUGACUGUCUCGUAAAUCU ......(((((.............((((......))))......((((((...(((.........)))....))))))...)))))((((...((......)).)))). (-23.00 = -23.40 + 0.40)



| Location | 13,867,194 – 13,867,312 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -25.87 |
| Energy contribution | -29.32 |
| Covariance contribution | 3.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13867194 118 + 22407834 GCUGAUUGAGUCUGGGCGAUUGUGACUGUCUCGUAAAUCUGCGAA--GAAGGUGUGAGCUGAGGGACAAAAGCCCAGACGCAUUUAAGCUAAUUUGUUCUGACGUCGCCUGCGACGUGAC (((...((.((((((((..((((..((.(((((((....)))).)--)).(((....))).))..))))..)))))))).))....)))............((((((....))))))... ( -41.90) >DroSec_CAF1 11262 118 + 1 GCUGAUUGAGUUUGGGCGAUUGCGACUGUCUCGUAAAUCUGCGAA--GAAGGUGUGAGCUGAGAGACAAAAGCCCAGACGCAUUUAAGCUAAUUUGUUCUGACGUCGCCUGCGACGUGAC (((...((.(((((((((....)...((((((.....(((....)--)).(((....)))..))))))...)))))))).))....)))............((((((....))))))... ( -36.90) >DroSim_CAF1 12773 118 + 1 GCUGAUUGAGUCUGGGCGAUUGUGACUGUCUCGUAAAUCUGCGAA--GAAGGUGUGAGCCGAGGGACAAAAGCCCAGACGCAUUUAAGCUAAUUUGUUCUGACGUCGCCUGCGACGUGAC (((...((.((((((((..((((..((.(((((((....)))).)--)).(((....))).))..))))..)))))))).))....)))............((((((....))))))... ( -44.50) >DroEre_CAF1 11562 118 + 1 GCUGAUUGAGUCUGAGCGAUUGUGACUGUCUCGUAAAUCAGAAGA--GGAGGUGCGAGAUCAGGGACAAAAGUCCAGACACAUUUAAGCUAAUUUGUUCUGACGUCGCCUGCGACGUGAC (((...((.(((((..(..((((..((((((((((..((......--.))..))))))).)))..))))..)..))))).))....)))............((((((....))))))... ( -43.60) >DroYak_CAF1 12929 118 + 1 GCUGAUUGAGUUUGGGCGAUUGUGACUGUCUCGUAAAUCUGGGAA--GAAGGUGUGAGAUCAGGGACAAAAGCCCAGACGCAUUUAAGCUAAUUUGUUCUGACGUCGCCAGCGACGUGAC (((...((.((((((((..((((..((((((((...((((.....--..)))).))))).)))..))))..)))))))).))....)))............((((((....))))))... ( -44.90) >DroAna_CAF1 10307 93 + 1 GCUGA----------------------GGCUCAUAACUCUGGGCUGGAAAGGUGUGAGAUGAAGGACAAAAGCCCGGCC-AACUUAAGCUAAUUUGUUCUGACA----CUCACACGUUGC .((..----------------------((((((......))))))....))(((((((.((.(((((((((((..((..-..))...)))..))))))))..))----)))))))..... ( -28.30) >consensus GCUGAUUGAGUCUGGGCGAUUGUGACUGUCUCGUAAAUCUGCGAA__GAAGGUGUGAGAUGAGGGACAAAAGCCCAGACGCAUUUAAGCUAAUUUGUUCUGACGUCGCCUGCGACGUGAC (((...((.((((((((..((((..((..((((...((((.........)))).))))...))..))))..)))))))).))....)))............((((((....))))))... (-25.87 = -29.32 + 3.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:46 2006