| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,851,564 – 13,851,661 |
| Length | 97 |
| Max. P | 0.615708 |

| Location | 13,851,564 – 13,851,661 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.09 |
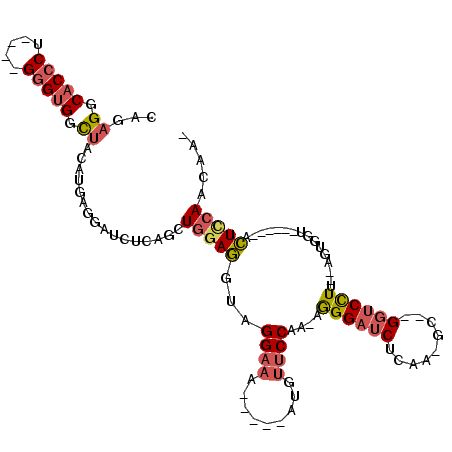
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -17.67 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.62 |
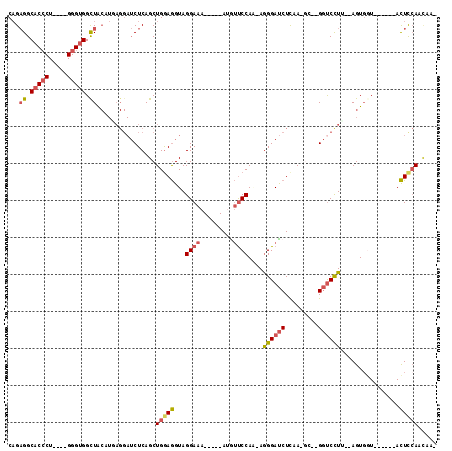
| Combinations/Pair | 1.23 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
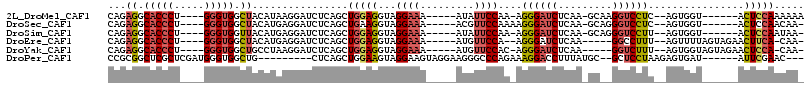
>2L_DroMel_CAF1 13851564 97 + 22407834 CAGAGGCACCCU----GGGUGGCUACAUAAGGAUCUCAGCUGGAGGUAGGAAA-----AUAUUCCAA-AGGGAUCUCAA-GCAAGGUCCUC--AGUGGU------ACUCCAAAAAA ..(((.((((..----.))))(((((...(((((((..(((.(((((.((((.-----...))))..-....))))).)-)).))))))).--.)))))------.)))....... ( -30.20) >DroSec_CAF1 4726 97 + 1 CAGAGGCACCCU----GGGUGGCUACAUGAGGAUCUCAGCUGAAGGUAGGAAA-----ACGUUCCAAAAGGGAUCUCAA-GCAGGGUCCUC--AGUGGU------ACUCCAACAA- ..(((.((((..----.))))(((((.(((((((((..(((..((((.((((.-----...)))).......))))..)-)).))))))))--))))))------.)))......- ( -32.80) >DroSim_CAF1 6288 96 + 1 CAGAGGCACCCU----GGGUGGUUACAUGAGGAUCUCAGCUGGAGGUAGGAAA-----AUAUUCCAA-AGGGAUCUCAA-GCAGGGUCCUU--AGUGGU------ACUCCAAUAA- ..((((.(((((----(.(((....)))(((.(((((...(((((.((.....-----.))))))).-.))))))))..-.))))))))))--..(((.------...)))....- ( -30.30) >DroEre_CAF1 4882 96 + 1 CAGAGGCACCCU----GGGUGGCUACAUGAGGAUCUCAGCUGGAGGUAGGAAA-----AUGUUCCA--AGGGAUCUCAA-----GGCCUUU--AGUUUUAGUAGAACUUCA-CAA- ...((.((((..----.)))).))...(((((.((((((((((((((..((..-----..((((..--..)))).))..-----.))))))--))))...).))).)))))-...- ( -26.80) >DroYak_CAF1 6266 97 + 1 CAGAGGCACCCU----GGGUGGCUGCCUAAGGAUCUCAGCUGGAGGUAGGAAA-----AUGUUCCAC-AGGGAUCUCAA-----GGUCUUU--AGUGGUAGUAGAACUCCA-CAA- ...((.((((..----.)))).))((((..(((((((.((.....)).((((.-----...))))..-.)))))))..)-----)))....--.((((.((.....)))))-)..- ( -29.90) >DroPer_CAF1 6855 96 + 1 CCGCGGCUCGCUCGAUGGGUGGCUG---------CUCAGCUGGAAGUAGGAAGUAGGAAGGGCCCAGAAAGGACCUUUAUGC--GCUCCUAAGAGUGAU------AUUCGAAC--- ..(((((.((((.....))))))))---------)(((.((.....(((((.(((.(((((..((.....)).))))).)))--..)))))..))))).------........--- ( -30.30) >consensus CAGAGGCACCCU____GGGUGGCUACAUGAGGAUCUCAGCUGGAGGUAGGAAA_____AUGUUCCAA_AGGGAUCUCAA_GC__GGUCCUU__AGUGGU______ACUCCAACAA_ ...((.(((((.....))))).))................(((((...((((.........))))....((((((.........))))))................)))))..... (-17.67 = -18.28 + 0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:44 2006