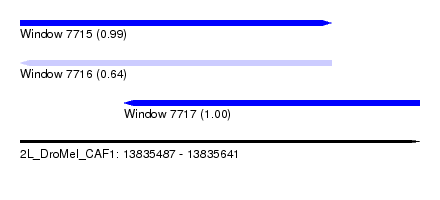
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,835,487 – 13,835,641 |
| Length | 154 |
| Max. P | 0.996796 |

| Location | 13,835,487 – 13,835,607 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -25.14 |
| Energy contribution | -26.18 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13835487 120 + 22407834 UUAGUUAAAAAUUAAUCACCCACUUGAGUUGGUGGAAGUUCGCUCUGUAGAGUGACCACUGUACCUGCUCUUCAUCCACAGUUUGUUGUGGAACGCCUCGCACCAACUACAUACUUUUAC ........................((((((((((((((...((...(((.(((....))).)))..)).)))).(((((((....)))))))........)))))))).))......... ( -31.60) >DroSec_CAF1 2063 120 + 1 CAACUUAAAAUUAAAUCACCCAUUUGAGUUGGUGGAAGUUCGCUCGGUAGAGUUACCACUGUACCUGCUCUUCUUCCACAGUUUGUUGUGGAACGCCUCGCACCAACUACAUACUUUUAC ........................((((((((((((((...((..((((.(((....))).)))).)).))))((((((((....)))))))).......)))))))).))......... ( -33.60) >DroSim_CAF1 3216 120 + 1 CAACUUAAAAUUAAAUCACCUAUUUGAGUUGGUGGAAGUUCGCUCUGUAGAGUUACCACUGUACCUGCUAUUCUUCCACAGUUUGUUGUGGAACGCCUCGCACCAACUACAUACUUUUAC (((((((((.............)))))))))(((((((...((...(((.(((....))).)))..))....)))))))(((.(((..(((..((...))..)))...))).)))..... ( -29.42) >DroEre_CAF1 3187 119 + 1 CCAAUUAAAAUUCAAUCAAAUAUUUGAAUUGGUG-AAGUUGGCUCUGUAGAGUGACCACUGUACCUGCUCUUCUUCCACAGUUUGUUGUGGAACGCCUCGCACCGACUACAUACUUUUAC ........(((((((........)))))))((((-.....(((...(((.(((....))).))).........((((((((....)))))))).)))...))))................ ( -28.80) >DroYak_CAF1 3041 120 + 1 CCAAUUAAAAUUGACCCACCCAUUUGAGUUGGUGGAAAUUCGCUCCAUAGAGCGAUCACUGUACCUGCUCUUCUUCCACAGUUUGUUGUGGAACGCCUCGCACCAACUACAUACUUUUAC .((((....))))...........((((((((((((...((((((....))))))..................((((((((....))))))))....)).)))))))).))......... ( -30.70) >consensus CAAAUUAAAAUUAAAUCACCCAUUUGAGUUGGUGGAAGUUCGCUCUGUAGAGUGACCACUGUACCUGCUCUUCUUCCACAGUUUGUUGUGGAACGCCUCGCACCAACUACAUACUUUUAC ........................((((((((((((((...((...(((.(((....))).)))..)).)))).(((((((....)))))))........)))))))).))......... (-25.14 = -26.18 + 1.04)

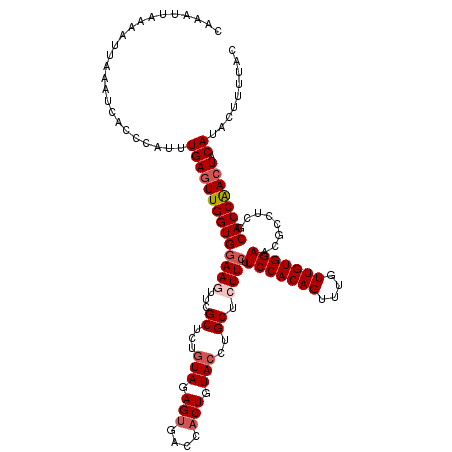
| Location | 13,835,487 – 13,835,607 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -27.74 |
| Energy contribution | -27.70 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13835487 120 - 22407834 GUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAUGAAGAGCAGGUACAGUGGUCACUCUACAGAGCGAACUUCCACCAACUCAAGUGGGUGAUUAAUUUUUAACUAA .........((.((((((((.((((...((((((......))))))......((..(((.(((....))).)))...))...))))))))))))))........................ ( -30.40) >DroSec_CAF1 2063 120 - 1 GUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAGAGCAGGUACAGUGGUAACUCUACCGAGCGAACUUCCACCAACUCAAAUGGGUGAUUUAAUUUUAAGUUG .........((.((((((((.((((..(((((((......))))))).....((.((((.(((....))).))))..))...)))))))))))))).......((((((....)))))). ( -35.80) >DroSim_CAF1 3216 120 - 1 GUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAUAGCAGGUACAGUGGUAACUCUACAGAGCGAACUUCCACCAACUCAAAUAGGUGAUUUAAUUUUAAGUUG .........((.((((((((.((((..(((((((......))))))).....((..(((.(((....))).)))...))...)))))))))))))).......((((((....)))))). ( -32.30) >DroEre_CAF1 3187 119 - 1 GUAAAAGUAUGUAGUCGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAGAGCAGGUACAGUGGUCACUCUACAGAGCCAACUU-CACCAAUUCAAAUAUUUGAUUGAAUUUUAAUUGG ................((((.(((((.(((((((......))))))).....)).(((.(.((((.....)))).).)))..)))-))))(((((((........)))))))........ ( -27.10) >DroYak_CAF1 3041 120 - 1 GUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAGAGCAGGUACAGUGAUCGCUCUAUGGAGCGAAUUUCCACCAACUCAAAUGGGUGGGUCAAUUUUAAUUGG .........((.((((((((.(((((.(((((((......))))))).....))...........((((((....))))))..)))))))))))))...........((((....)))). ( -33.90) >consensus GUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAGAGCAGGUACAGUGGUCACUCUACAGAGCGAACUUCCACCAACUCAAAUGGGUGAUUUAAUUUUAAGUGG .........((.((((((((.((((..(((((((......))))))).....((..(((.(((....))).)))...))...))))))))))))))........................ (-27.74 = -27.70 + -0.04)

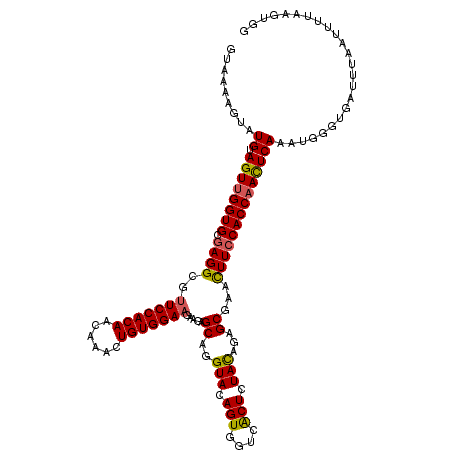
| Location | 13,835,527 – 13,835,641 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -32.58 |
| Energy contribution | -32.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13835527 114 - 22407834 AGUGCGAGAAUUUACGUCGAGCGCAGCCGAGUAUGUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAUGAAGAGCAGGUACAGUGGUCACUCUACAGAGCG .((((........(((((...((((.((((.((((((....)))))).)))))))).)))))((((((......))))))..........)))).((((.....))))...... ( -35.70) >DroSec_CAF1 2103 114 - 1 AGUGCGAGAAUUUACGUCGAGCCCAGCCGAGUAUGUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAGAGCAGGUACAGUGGUAACUCUACCGAGCG ..(((.........((((..(((((((...((((......))))...))))).))..))))(((((((......))))))).....)))((((.(((....))).))))..... ( -36.50) >DroSim_CAF1 3256 114 - 1 AGUGCGAGAAUUUACGUCGAGCGCAGCCGAGUAUGUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAUAGCAGGUACAGUGGUAACUCUACAGAGCG .((((.........((((...((((.((((.((((((....)))))).)))))))).))))(((((((......))))))).........)))).((((.....))))...... ( -35.90) >DroEre_CAF1 3226 114 - 1 AGUGCGAGAAUUUACGUCGAGCGCAGCCGAGUAUGUAAAAGUAUGUAGUCGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAGAGCAGGUACAGUGGUCACUCUACAGAGCC ..(((.........((((...((((.((((.((((((....)))))).)))))))).))))(((((((......))))))).....)))(((.(.((((.....)))).).))) ( -38.50) >DroYak_CAF1 3081 114 - 1 AGUGCGAGAAUUUACGUCGAGCGCAGACGAGUAUGUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAGAGCAGGUACAGUGAUCGCUCUAUGGAGCG .((((.........((((..((((.(((..((((......))))...))).))))..))))(((((((......))))))).........))))......(((((....))))) ( -36.50) >consensus AGUGCGAGAAUUUACGUCGAGCGCAGCCGAGUAUGUAAAAGUAUGUAGUUGGUGCGAGGCGUUCCACAACAAACUGUGGAAGAAGAGCAGGUACAGUGGUCACUCUACAGAGCG ..(((........(((((...((((.((((.((((((....)))))).)))))))).)))))((((((......))))))......))).(((.(((....))).)))...... (-32.58 = -32.50 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:40 2006