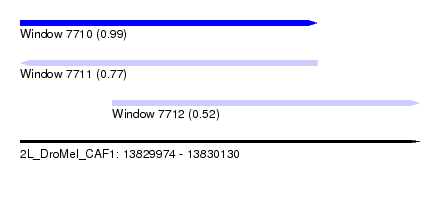
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,829,974 – 13,830,130 |
| Length | 156 |
| Max. P | 0.992539 |

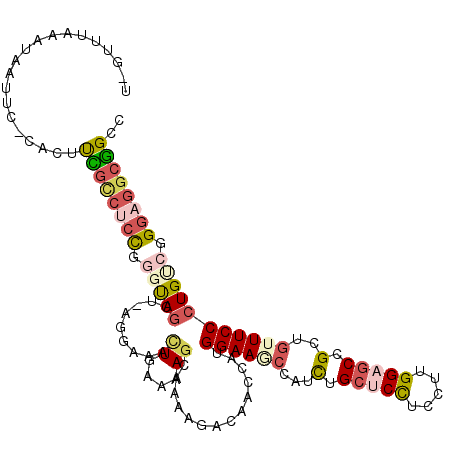
| Location | 13,829,974 – 13,830,090 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.50 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -21.04 |
| Energy contribution | -22.85 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13829974 116 + 22407834 CCGUUUAAAUAACAC-CACUUCGCCUGUGGGUAGCUGAGGAAACUUAAAAGCAGACGACAGCUAUGGAAGCCAUCUGCUCCUCCUUGGAGCCGCUGUUUCCCUGUCGGGAGGCGGCC .(((((.....((.(-(((.......)))))).((((((....)))...))))))))...(((.....))).....(((((.....))))).((((((((((....)))))))))). ( -42.30) >DroPse_CAF1 4809 96 + 1 ------------UGCGCGCUUUAUCCCCG-GCAGC-----UG-UUUGUAAGCAAAUCGC--AAAUGGAAAAUAUCUGCGCUGCGUUGGAGCCGCUGUUUCCCUGCCGGGAGGGAGCA ------------.....(((((...((((-(((((-----.(-((((....))))).))--....(((((......(((..((......)))))..))))).)))))))..))))). ( -35.40) >DroSec_CAF1 1757 116 + 1 UUGUUUAAAUAAUUC-CACUUCGCCCCCGGCUAGCUGAGAAAACUGAAAAGCGGAAGGCAACCAUGGAAGCCAUCUGCUCCUCCUUGGAGCCGCUGUUUCCCUGUCGGGAGGCGGCC ...............-....(((((((((((.((..((((.........((((((.(((..(....)..))).)).(((((.....))))))))).)))).)))))))).))))).. ( -40.40) >DroSim_CAF1 1756 103 + 1 ------------UUC-CACUUCGCCUCCGGGUAGCU-AGGAAACUAAAAAACGGAAGGCAACCAUGGAAGCCAUCUGCUCCUCCUUGGAACCGCUGUUUCCCUGUCGGGAGGCGGCC ------------...-......(((((((..(...(-((....)))..)..))).))))..(((.((((((.....)))..))).)))....((((((((((....)))))))))). ( -40.60) >DroEre_CAF1 1746 116 + 1 UCGUUUAAAUAAUUC-UAUCCUGCCUCUGGGCAGAUCCGUAAUCUGAAAAGAAAAAAACACCAAUGGAAUCCAUUUGCUCCUCCUUGGCGCCGCUGUUUCCCUGUCGGGAAACGGCU ......((((.((((-(((.(((((....)))))........(((....)))...........)))))))..))))((.((.....)).)).((((((((((....)))))))))). ( -36.00) >DroYak_CAF1 1745 113 + 1 UUGUUUAAAUAAUUG-AUCUUUGCCUCCG--UAAUUCAGUACUCUGAAAAGAGA-AAACACCCAUGGAAUCCAUCUGCUCCUCCUUGGAGCCGAUGUUUCCCUGUCGGGAGGCGGCC ...............-....((((((((.--..........((((....)))).-.......((.((((..((((.(((((.....))))).)))).)))).))...)))))))).. ( -38.00) >consensus U_GUUUAAAUAAUUC_CACUUCGCCUCCGGGUAGCU_AGGAAACUGAAAAGCAAAAGACAACCAUGGAAGCCAUCUGCUCCUCCUUGGAGCCGCUGUUUCCCUGUCGGGAGGCGGCC ....................((((((((.(((((...((....))....................((((((...(.(((((.....))))).)..))))))))))).)))))))).. (-21.04 = -22.85 + 1.81)

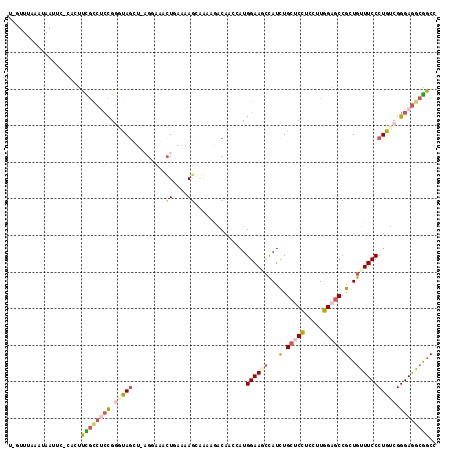
| Location | 13,829,974 – 13,830,090 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.50 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13829974 116 - 22407834 GGCCGCCUCCCGACAGGGAAACAGCGGCUCCAAGGAGGAGCAGAUGGCUUCCAUAGCUGUCGUCUGCUUUUAAGUUUCCUCAGCUACCCACAGGCGAAGUG-GUGUUAUUUAAACGG ((((((.((((....))))....))))))((....(((((((((((((..........)))))))))))))..((((....(((...((((.......)))-).)))....)))))) ( -40.50) >DroPse_CAF1 4809 96 - 1 UGCUCCCUCCCGGCAGGGAAACAGCGGCUCCAACGCAGCGCAGAUAUUUUCCAUUU--GCGAUUUGCUUACAAA-CA-----GCUGC-CGGGGAUAAAGCGCGCA------------ .(((...((((((((((....).(((.......)))..(((((((.......))))--))).............-..-----.))))-)))))....))).....------------ ( -33.30) >DroSec_CAF1 1757 116 - 1 GGCCGCCUCCCGACAGGGAAACAGCGGCUCCAAGGAGGAGCAGAUGGCUUCCAUGGUUGCCUUCCGCUUUUCAGUUUUCUCAGCUAGCCGGGGGCGAAGUG-GAAUUAUUUAAACAA .(((((.((((....))))....)).(((((.....)))))....)))((((((..((((((((.(((....((((.....))))))).)))))))).)))-)))............ ( -42.90) >DroSim_CAF1 1756 103 - 1 GGCCGCCUCCCGACAGGGAAACAGCGGUUCCAAGGAGGAGCAGAUGGCUUCCAUGGUUGCCUUCCGUUUUUUAGUUUCCU-AGCUACCCGGAGGCGAAGUG-GAA------------ ((((((.((((....))))....)).(((((.....)))))....))))(((((..((((((.(((.....(((((....-)))))..))))))))).)))-)).------------ ( -39.80) >DroEre_CAF1 1746 116 - 1 AGCCGUUUCCCGACAGGGAAACAGCGGCGCCAAGGAGGAGCAAAUGGAUUCCAUUGGUGUUUUUUUCUUUUCAGAUUACGGAUCUGCCCAGAGGCAGGAUA-GAAUUAUUUAAACGA .((.(((((((....))))))).))((((((((((((...(.....).)))).))))))))...((((.((((((((...)))))(((....))).))).)-)))............ ( -36.10) >DroYak_CAF1 1745 113 - 1 GGCCGCCUCCCGACAGGGAAACAUCGGCUCCAAGGAGGAGCAGAUGGAUUCCAUGGGUGUUU-UCUCUUUUCAGAGUACUGAAUUA--CGGAGGCAAAGAU-CAAUUAUUUAAACAA ....((((((.....((((((((((.(((((.....)))))..((((...)))).)))))))-)))...(((((....)))))...--.))))))......-............... ( -37.40) >consensus GGCCGCCUCCCGACAGGGAAACAGCGGCUCCAAGGAGGAGCAGAUGGCUUCCAUGGGUGCCUUCCGCUUUUCAGUUUACU_AGCUACCCGGAGGCAAAGUG_GAAUUAUUUAAAC_A .(((.((((((....))))..((((.(((((.....)))))..(((.....))).))))..............................)).)))...................... (-18.72 = -19.58 + 0.86)

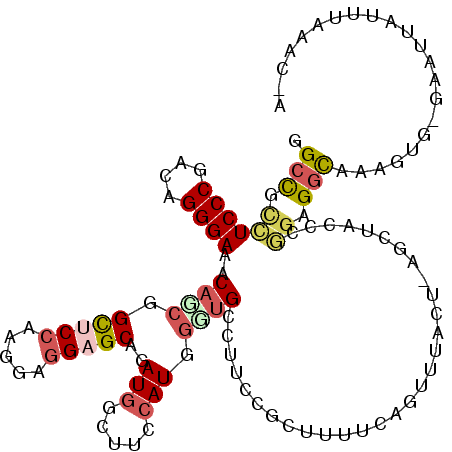
| Location | 13,830,010 – 13,830,130 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -25.72 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13830010 120 + 22407834 AGGAAACUUAAAAGCAGACGACAGCUAUGGAAGCCAUCUGCUCCUCCUUGGAGCCGCUGUUUCCCUGUCGGGAGGCGGCCAUUGAAACGCUGGGCGAGCUGAUAGGAGAUUCGAGUGAGA ((....))....((((((.(...(((.....)))).)))))).(((((((((.((((((((((((....)))))))))).........(((.....))).....))...)))))).))). ( -40.50) >DroSec_CAF1 1793 120 + 1 AGAAAACUGAAAAGCGGAAGGCAACCAUGGAAGCCAUCUGCUCCUCCUUGGAGCCGCUGUUUCCCUGUCGGGAGGCGGCCAUUGAAACGCUGGGCGAGCUAAUAGGAGAUUCCAGUGAGG .....((((...((((((.(((..(....)..))).)))))).(((((...(((.((((((((((....))))))))))........(((...))).)))...)))))....)))).... ( -47.10) >DroSim_CAF1 1779 120 + 1 AGGAAACUAAAAAACGGAAGGCAACCAUGGAAGCCAUCUGCUCCUCCUUGGAACCGCUGUUUCCCUGUCGGGAGGCGGCCAUUGAAACGCUGGGCGAGCUGAUAGGAGAUUCCAGUGAGG ((....))......((((.(((..(....)..))).))))..((((.((((((((((((((((((....)))))))))).........(((.....))).....))...)))))).)))) ( -42.80) >DroEre_CAF1 1782 120 + 1 CGUAAUCUGAAAAGAAAAAAACACCAAUGGAAUCCAUUUGCUCCUCCUUGGCGCCGCUGUUUCCCUGUCGGGAAACGGCUAUUGAAACGCUCGGCGAGCUGAUAGGAGAUUCCCGUGAAG .....(((....)))......(((....((((((.......((((..(..(((((((((((((((....))))))))))....((.....)))))..))..).)))))))))).)))... ( -38.51) >DroYak_CAF1 1779 119 + 1 AGUACUCUGAAAAGAGA-AAACACCCAUGGAAUCCAUCUGCUCCUCCUUGGAGCCGAUGUUUCCCUGUCGGGAGGCGGCCAUUGAAACGCUUGGCGAGCUGAUAGGAGAUUCCAGUGAGG ....((((....)))).-.....(((((((((((((((.(((((.....))))).))))....((((((....(((.((((..........))))..))))))))).))))))).)).)) ( -46.10) >DroPer_CAF1 4265 114 + 1 ---UG-UUUGUAAGCAAAUCGC--AAAUGGAAAAUAUCUGCGCUGCGUUGGAGCCGCUGUUUCCCUGCCGGGAGGGAGCAAUAAAAACACUGGGCGAGCUGAUUGGCGACUCCCGGGAAG ---((-(((....(((...(((--(.(((.....))).)))).))).........(((.((((((....)))))).))).....)))))(((((.(.(((....)))..).))))).... ( -34.70) >consensus AGGAAACUGAAAAGCAAAAGACAACCAUGGAAGCCAUCUGCUCCUCCUUGGAGCCGCUGUUUCCCUGUCGGGAGGCGGCCAUUGAAACGCUGGGCGAGCUGAUAGGAGAUUCCAGUGAGG ............................((((.(((((.(((((((...(....)((((((((((....))))))))))............))).)))).))).))...))))....... (-25.72 = -26.58 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:34 2006