
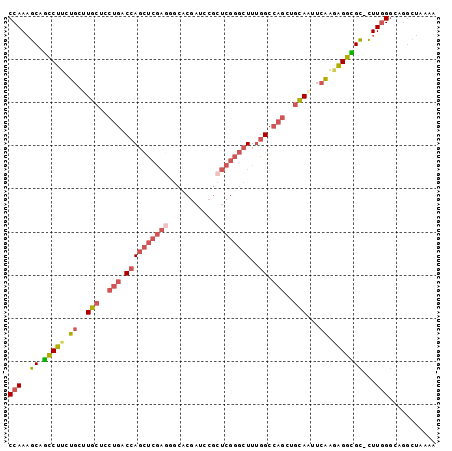
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,813,011 – 13,813,109 |
| Length | 98 |
| Max. P | 0.873052 |

| Location | 13,813,011 – 13,813,109 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -22.84 |
| Energy contribution | -24.15 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13813011 98 + 22407834 CCAAAGCAGCCUUCUCCUUGCUCCUGACCAGCUCGAGUGCACGAUUCGCUCGGGCUUUGGCCAGCUGCAAUUCGAGAGGCGC-CUUUGGCAGGUUAAAA ((((((..((((.(((.((((..(((.((((((((((((.......)))))))))..))).)))..))))...)))))))..-)))))).......... ( -41.50) >DroSec_CAF1 960 98 + 1 CCAAAGCAGCCUUCUGCUUGCUCCUGACCAGCUCGAGCUCCCGAUCCGCUCGGGCUUUGGCCAGCUGCAAUUCGAGAGGCGC-CUUGGGCAGGUUAAAA ((((.((.((((..((.((((..(((.(((((((((((.........))))))))..))).)))..))))..))..))))))-.))))........... ( -40.20) >DroSim_CAF1 960 99 + 1 CCAAAGCAGCCUUCUGCUUGCUCCUGACCAGCUCGAGCUCCCGAUCCGCUCGGGCUUUGGCCAGCUGCAAUUCGAGAGGCGCCCUUGGGCAGGUUAAAA ((......((((..((.((((..(((.(((((((((((.........))))))))..))).)))..))))..))..))))(((....))).))...... ( -40.80) >DroEre_CAF1 963 98 + 1 CCAAAGCAGCCUUCUGCUUGCUCCUGACCAGCUCGAUGGCCCGAUCCACUCGGGCUUUGGCCAGCUGCACUUCAAAAGGUGC-CUUGGGAACCCUAUAA ((((.(((.((((.((..(((..(((.((((......(((((((.....))))))))))).)))..)))...)).)))))))-.))))........... ( -35.30) >DroYak_CAF1 961 98 + 1 CCAAAGCAGCCUUCUGCGUGCUCCUGACCAGCUCGAGGGCACGAUCCGCUCGGGCUUUGGCCAGCUGCACUUCAAAGGGCGC-CUUGGGCUGGCUGAAA ((((.((.(((((.((.((((..(((.((((((((((((......)).)))))))..))).)))..))))..)).)))))))-.))))........... ( -41.20) >DroAna_CAF1 942 98 + 1 CCAAUGCCUUCUUUUGGUUGCUUGAGACCAUUUGACUGGCACGACUGGCCCGUUCUUCAGCAAGCUUUAUCUCAGUAGGAGU-UUUGGACAGGCUCCAA ....((((.((...(((((......)))))...))..)))).....((((.((((......(((((((.........)))))-)).)))).)))).... ( -22.90) >consensus CCAAAGCAGCCUUCUGCUUGCUCCUGACCAGCUCGAGGGCACGAUCCGCUCGGGCUUUGGCCAGCUGCAAUUCAAGAGGCGC_CUUGGGCAGGCUAAAA (((..((.(((((.((..(((..(((.((((((((((...........))))))))..)).)))..)))...)).)))))))...)))........... (-22.84 = -24.15 + 1.31)


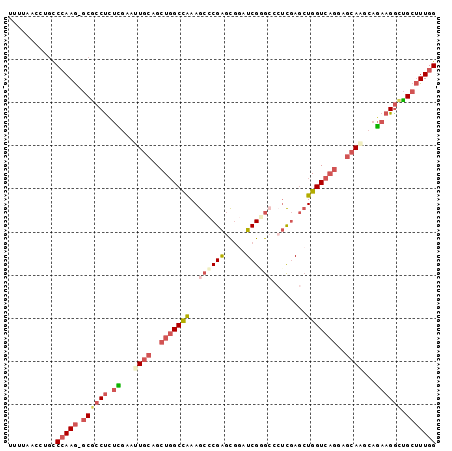
| Location | 13,813,011 – 13,813,109 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -22.81 |
| Energy contribution | -24.62 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13813011 98 - 22407834 UUUUAACCUGCCAAAG-GCGCCUCUCGAAUUGCAGCUGGCCAAAGCCCGAGCGAAUCGUGCACUCGAGCUGGUCAGGAGCAAGGAGAAGGCUGCUUUGG ..........((((((-..(((((((...((((..(((((((..((.((((.(.......).)))).)))))))))..)))).))).))))..)))))) ( -38.90) >DroSec_CAF1 960 98 - 1 UUUUAACCUGCCCAAG-GCGCCUCUCGAAUUGCAGCUGGCCAAAGCCCGAGCGGAUCGGGAGCUCGAGCUGGUCAGGAGCAAGCAGAAGGCUGCUUUGG ...........(((((-((((((.((...((((..(((((((..((.(((((.........))))).)))))))))..))))...)))))).))))))) ( -44.00) >DroSim_CAF1 960 99 - 1 UUUUAACCUGCCCAAGGGCGCCUCUCGAAUUGCAGCUGGCCAAAGCCCGAGCGGAUCGGGAGCUCGAGCUGGUCAGGAGCAAGCAGAAGGCUGCUUUGG ...........((((((..((((.((...((((..(((((((..((.(((((.........))))).)))))))))..))))...))))))..)))))) ( -40.30) >DroEre_CAF1 963 98 - 1 UUAUAGGGUUCCCAAG-GCACCUUUUGAAGUGCAGCUGGCCAAAGCCCGAGUGGAUCGGGCCAUCGAGCUGGUCAGGAGCAAGCAGAAGGCUGCUUUGG ...........(((((-((((((((((...(((..(((((((..((((((.....))))))........)))))))..)))..))))))).)))))))) ( -44.90) >DroYak_CAF1 961 98 - 1 UUUCAGCCAGCCCAAG-GCGCCCUUUGAAGUGCAGCUGGCCAAAGCCCGAGCGGAUCGUGCCCUCGAGCUGGUCAGGAGCACGCAGAAGGCUGCUUUGG ...........(((((-(((.(((((...((((..(((((((..((.((((.((......)))))).)))))))))..))))...))))).)))))))) ( -45.80) >DroAna_CAF1 942 98 - 1 UUGGAGCCUGUCCAAA-ACUCCUACUGAGAUAAAGCUUGCUGAAGAACGGGCCAGUCGUGCCAGUCAAAUGGUCUCAAGCAACCAAAAGAAGGCAUUGG ...(.((((((.(...-.(((.....)))....((....))...).)))))))....(((((..((...((((........))))...)).)))))... ( -20.10) >consensus UUUUAACCUGCCCAAG_GCGCCUCUCGAAUUGCAGCUGGCCAAAGCCCGAGCGGAUCGGGCCCUCGAGCUGGUCAGGAGCAAGCAGAAGGCUGCUUUGG ...........(((((.((((((.((...((((..(((((((..((((((.....))))))........)))))))..))))...)))))).))))))) (-22.81 = -24.62 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:29 2006