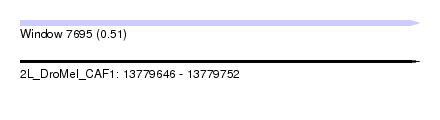
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,779,646 – 13,779,752 |
| Length | 106 |
| Max. P | 0.505199 |

| Location | 13,779,646 – 13,779,752 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.67 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13779646 106 + 22407834 UGCAAGUUGAUGGUGCAGGUUGUAUUGUUUCCGCGAUCCAGAACAACUACACUCGUUGCGGCCAUUGCGGCCGCUGUCGUGGCCAUUUU--GAAUU-UUC-----CUAACGGUA ..(((...((((((...((((((.(((..((...))..))).)))))).(((.....((((((.....))))))....)))))))))))--)....-..(-----(....)).. ( -31.70) >DroVir_CAF1 9729 105 + 1 UGCAAGUUGAUUGUGCAUGUCGUAUUAUUGCCACGAUCCAAUACGACGACACUGGUGGCUGCCACGGCAGCCGCUGUCGUGGCCAUUUU--GUUU-CGAU-----GCAACGGU- ((((.(..(((.((((.....)))).)))..).(((..(((..(.((((((...((((((((....)))))))))))))).).....))--)..)-)).)-----))).....- ( -36.90) >DroPse_CAF1 8394 108 + 1 UGCAAGUUGAUGGUGCAGGUCGUGUUAUUGCCGCGAUCCAAAACGACGACACUGGUGGCGGCAAUGGCGGCCGCUGUCGUGGCCAUAUU--UGUUU-UUU--AGACUAACGGU- ((((.........))))((((..(((((((((((.(.(((............)))).)))))))))))((((((....)))))).....--.....-...--.))))......- ( -38.90) >DroGri_CAF1 9439 106 + 1 UGCAAGUUGAUUGUGCAUGUCGUAUUAUUGCCACGAUCCAAUACAACAACACUGGUCGCUGCCACGGCAGCCGCAGUCGUGGCCAUUUU--GUUUUUGGG-----GCAACGGU- (((..((((((((((((...........)).)))))).)))).(((.((((.((((((((((....))))).((....)))))))...)--))).)))..-----))).....- ( -29.70) >DroMoj_CAF1 10318 105 + 1 UGCAAGUUGAUUGUGCAUGUCGUAUUAUUGCCACGAUCCAAUACAACAACACUGGUGGCUGCUACGGCAGCCGCAGUCGUGGCCAUUUU--GCUU-ACAC-----GUAACGGU- .........(((((((.(((.(((..((.((((((((.........((....))((((((((....)))))))).)))))))).))..)--))..-))).-----)).)))))- ( -37.90) >DroAna_CAF1 8267 112 + 1 UGCAAGUUAAUGGUGCAGGUUGUGUUGUUGCCGCGAUCCAAUACCACAACGCUGGUGGCGGCCAUUGCGGCCGCUGUCGUGGCCAUUUUUUAAAAU-UUCGGCGACUAACGGU- .....((((......(((((((((.(((((........))))).)))))).)))..(((((((.....)))))))(((((.(..((((....))))-..).)))))))))...- ( -37.40) >consensus UGCAAGUUGAUGGUGCAGGUCGUAUUAUUGCCACGAUCCAAUACAACAACACUGGUGGCGGCCACGGCAGCCGCUGUCGUGGCCAUUUU__GAUUU_UUC_____CUAACGGU_ .....((((((..(((.....)))..)).((((((((.........((....))(((((.((....)).))))).)))))))).......................)))).... (-23.64 = -23.67 + 0.03)

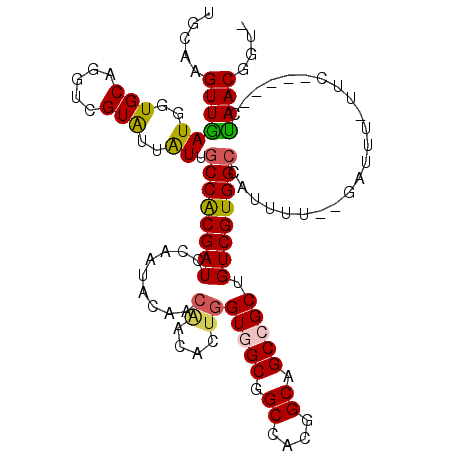
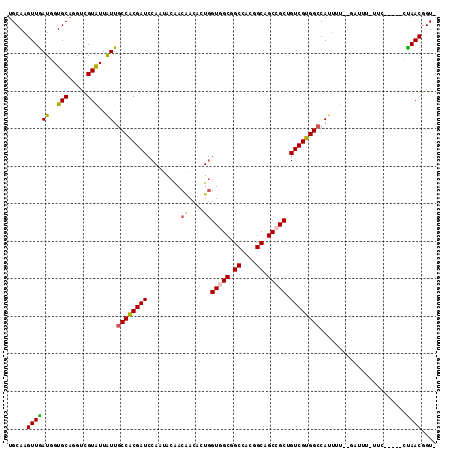
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:18 2006