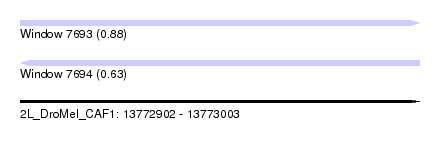
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,772,902 – 13,773,003 |
| Length | 101 |
| Max. P | 0.882337 |

| Location | 13,772,902 – 13,773,003 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -18.94 |
| Energy contribution | -18.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
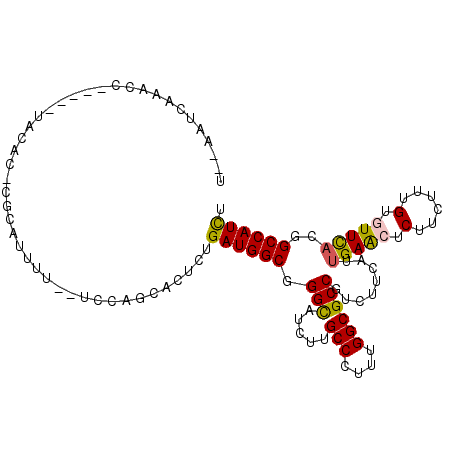
>2L_DroMel_CAF1 13772902 101 + 22407834 U--AAUCAAACCU----UCCAUGCGCAUUUU--UCCAGCACUCUGAUGGCGGGCAUCUUGCCCUUUGGCGCCGUCUUCAUUGAACUCUUCUUUGUGUUCACGGCCAUCU .--..........----....(((.......--....)))....((((((((((.....)))).................(((((.(......).)))))..)))))). ( -24.40) >DroGri_CAF1 1565 105 + 1 UAUACUCAACUC-GCUCUAAUC---UUUCUGUCCACAGCACUCUGAUGGCUGGCAUUUUGCCCUUUGGCGCCGUUUUUAUAGAGCUCUUCUUUGUCUUUACGGCCAUUU ............-(((......---.....(.((((((....))).)))).(((.....)))....)))(((((....((((((.....))))))....)))))..... ( -20.80) >DroSim_CAF1 1567 100 + 1 U--AAUCAAACU-----UUCACGCGCAUUUU--UGCAGCACUCUGAUGGCGGGCAUCUUGCCCUUUGGCGCCGUCUUCAUUGAACUCUUCUUUGUGUUCACGGCCAUCU .--.........-----.....(((((....--))).)).....((((((((((.....)))).................(((((.(......).)))))..)))))). ( -28.00) >DroEre_CAF1 1776 99 + 1 U--UAUCAAACU-----CAUAC-CGAAUUUU--UGCAGCACUCUGAUGGCUGGCAUCUUGCCCUUUGGCGCCGUCUUCAUUGAACUCUUCUUUGUGUUCACGGCCAUCU .--.........-----.....-........--...........((((((((.......(((....)))...........(((((.(......).))))))))))))). ( -22.50) >DroYak_CAF1 1564 99 + 1 U--AAUCAAACC-----UCCAC-CGCUUUUU--UCCAGCACUCUGAUGGCGGGUAUCUUGCCCUUUGGCGCCGUCUUCAUUGAACUCUUCUUUGUGUUCACGGCCAUCU .--.........-----.....-.(((....--...))).....((((((((((.....)))).................(((((.(......).)))))..)))))). ( -22.50) >DroAna_CAF1 1544 90 + 1 U--AAU---------------UGCAUAUCUU--UUUAGCACUUUGAUGGCUGGCAUUUUGCCGUUUGGCGCCGUCUUCAUUGAGCUCUUCUUUGUAUUCACGGCCAUCU .--...---------------(((.((....--..)))))....((((((((.......(((....)))((((.......)).))...............)))))))). ( -20.10) >consensus U__AAUCAAACC_____UACAC_CGCAUUUU__UCCAGCACUCUGAUGGCGGGCAUCUUGCCCUUUGGCGCCGUCUUCAUUGAACUCUUCUUUGUGUUCACGGCCAUCU ............................................((((((.(((.....(((....))))))........(((((.(......).)))))..)))))). (-18.94 = -18.80 + -0.14)

| Location | 13,772,902 – 13,773,003 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -16.53 |
| Energy contribution | -17.58 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
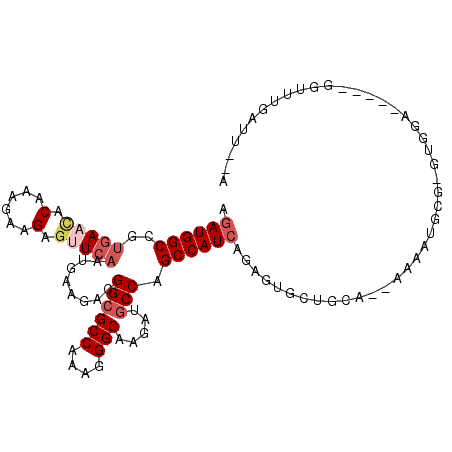
>2L_DroMel_CAF1 13772902 101 - 22407834 AGAUGGCCGUGAACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCCGCCAUCAGAGUGCUGGA--AAAAUGCGCAUGGA----AGGUUUGAUU--A .((((((..(((((.(......).))))).................((((.....))))))))))...((((....--.....)))).....----..........--. ( -27.40) >DroGri_CAF1 1565 105 - 1 AAAUGGCCGUAAAGACAAAGAAGAGCUCUAUAAAAACGGCGCCAAAGGGCAAAAUGCCAGCCAUCAGAGUGCUGUGGACAGAAA---GAUUAGAGC-GAGUUGAGUAUA ...((((...........(((.....)))........((((((....))).....))).))))((((..((((.(((.(.....---).))).)))-)..))))..... ( -21.70) >DroSim_CAF1 1567 100 - 1 AGAUGGCCGUGAACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCCGCCAUCAGAGUGCUGCA--AAAAUGCGCGUGAA-----AGUUUGAUU--A .((((((..(((((.(......).))))).................((((.....))))))))))...((((....--.....)))).....-----.........--. ( -27.40) >DroEre_CAF1 1776 99 - 1 AGAUGGCCGUGAACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCAGCCAUCAGAGUGCUGCA--AAAAUUCG-GUAUG-----AGUUUGAUA--A .((((((..(((((.(......).)))))........((((((....))).....))).))))))...((((((..--......))-)))).-----.........--. ( -25.40) >DroYak_CAF1 1564 99 - 1 AGAUGGCCGUGAACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUACCCGCCAUCAGAGUGCUGGA--AAAAAGCG-GUGGA-----GGUUUGAUU--A .....(((((((((.(......).)))).......)))))(((....))).((((..(((((((....))(((...--....))))-)))).-----.))))....--. ( -23.71) >DroAna_CAF1 1544 90 - 1 AGAUGGCCGUGAAUACAAAGAAGAGCUCAAUGAAGACGGCGCCAAACGGCAAAAUGCCAGCCAUCAAAGUGCUAAA--AAGAUAUGCA---------------AUU--A .((((((............((.....)).........((((((....))).....))).))))))....(((....--.......)))---------------...--. ( -18.50) >consensus AGAUGGCCGUGAACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCAGCCAUCAGAGUGCUGCA__AAAAUGCG_GUGGA_____GGUUUGAUU__A .((((((..(((((.(......).)))))........((((((....))).....))).))))))............................................ (-16.53 = -17.58 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:17 2006