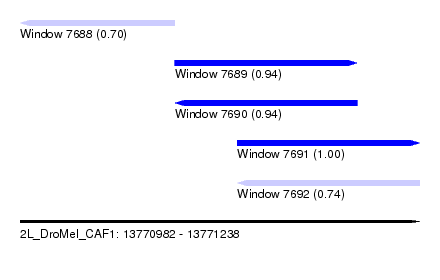
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,770,982 – 13,771,238 |
| Length | 256 |
| Max. P | 0.999350 |

| Location | 13,770,982 – 13,771,081 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -12.36 |
| Energy contribution | -13.16 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13770982 99 - 22407834 AAGUCGUGGCCUUAGGUAUGGUCAUACCAAUUAUCGACUAUCGAUAAGAGGCAAUACCGAUUGUUUUGAGAGAAACCGCGAAUAUUUAAGUACCGA--AAA----- ...((((((((((.(((((....)))))..((((((.....)))))))))))..........(((((....))))))))))...............--...----- ( -28.30) >DroSec_CAF1 33642 92 - 1 AAGUCGUGGCCAUCGGUAUGGUCACACCAAUUAU-------CGAUAACCAGCAAUACCGAUUGUAUUGAGAGGAACCGCGAAUAUUUAAACAUCGG--AAA----- ...((((((..((((((((((.....)))..)))-------))))..((..((((((.....))))))...))..))))))...............--...----- ( -23.80) >DroSim_CAF1 36665 92 - 1 AAGUCGUGGCCAUCGGUAUGGUCAUACCAAUUAU-------CGAUAACCAGCAAUACCGAUUGUAUUGAGAGGAACCGCAAAUAUUUAAACAUCGG--AAG----- .....((((..((((((((((.....)))..)))-------))))..((..((((((.....))))))...))..)))).................--...----- ( -20.50) >DroEre_CAF1 36965 105 - 1 AAGUCGUGGCCGAUGAAAAGGUCAUACUAAAUAUCGACUAUCGAUAAAUGGGAAUAUCGAUUGUAUUCAAAGUAACCGCAAAUAUUUAAUAUUU-UUGGAAUUUCU .(((.((((((........)))))))))...(((((.....)))))...((((((.((((..(((((..(((((........))))))))))..-)))).)))))) ( -21.80) >DroYak_CAF1 36694 101 - 1 AAGUCGUGGCCGUUGGUACGGUCAUACCAAUUAUCGACUAUCGAUGAAAGGGAAUAUCGAUUGUAUGCAAAGUAACCGCGAAUAUUAAAUAAUCGUUAGAA----- ...((((((...((((((((((((((((..((((((.....))))))...))..))).)))))))).))).....))))))....................----- ( -24.40) >consensus AAGUCGUGGCCAUCGGUAUGGUCAUACCAAUUAUCGACUAUCGAUAAAAGGCAAUACCGAUUGUAUUGAGAGGAACCGCGAAUAUUUAAAAAUCGG__AAA_____ .....((((((((....)))))))).....((((((.....))))))....((((((.....))))))...................................... (-12.36 = -13.16 + 0.80)

| Location | 13,771,081 – 13,771,198 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -27.77 |
| Energy contribution | -28.15 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13771081 117 + 22407834 ACCCAGCACCGAAACUCGGAAGAUUUGAAGAAGAAGAAUUGCCAGCAGGGAGACGCAGAAACUACAUUAUAUAUACAAAAUAUAUAGUUUCUAGCGGAAUUUCUGCCAACAAAAGGG .(((.((.(((.....)))..(((((.........)))))))..((((..(..(((((((((((...(((((.......))))))))))))).)))...)..))))........))) ( -31.70) >DroSec_CAF1 33734 117 + 1 ACCCAGCACCAAAACUCGGAAGAUUUGAAGAAGAAGAAUUGCCAGCAGGGAGACGCAGAAACUACAUUAUAUAUACAAAAUAUAUAGUUUCUAGCGUAAUUGCUGCCAACACAAGGG .(((.....((((..((....))))))...........(((.((((((....((((((((((((...(((((.......))))))))))))).))))..)))))).))).....))) ( -32.30) >DroSim_CAF1 36757 117 + 1 ACCCAGCACCAAAACUCGGAAGAUUUGAAGAAGAAGAAUUGCCAGCAGGGAGACGCAGAAACUACAUUAUAUAUACAAAAUAUAUAGUUUCUAGCGUAAUUGCUGCCAACACAAGGG .(((.....((((..((....))))))...........(((.((((((....((((((((((((...(((((.......))))))))))))).))))..)))))).))).....))) ( -32.30) >DroYak_CAF1 36795 117 + 1 ACUCAGCACCAAAACUCGGAAGAUUUGAAGAAGAAGAAUUGCCAGCAGGGAGACGCAGAAACUACAUUAUAUAUACCAAAUAUAUAGUUUCUUGCGGACUUGCUGCCAAAACAAGGG .(((.....((((..((....))))))...........(((.(((((((....(((((((((((...(((((.......)))))))))))).))))..))))))).))).....))) ( -30.90) >consensus ACCCAGCACCAAAACUCGGAAGAUUUGAAGAAGAAGAAUUGCCAGCAGGGAGACGCAGAAACUACAUUAUAUAUACAAAAUAUAUAGUUUCUAGCGGAAUUGCUGCCAACACAAGGG .(((.....((((..((....))))))...........(((.((((((....((((((((((((...(((((.......))))))))))))).))))..)))))).))).....))) (-27.77 = -28.15 + 0.38)


| Location | 13,771,081 – 13,771,198 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -28.09 |
| Energy contribution | -29.09 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13771081 117 - 22407834 CCCUUUUGUUGGCAGAAAUUCCGCUAGAAACUAUAUAUUUUGUAUAUAUAAUGUAGUUUCUGCGUCUCCCUGCUGGCAAUUCUUCUUCUUCAAAUCUUCCGAGUUUCGGUGCUGGGU (((..((((..((((......(((.(((((((((((..............)))))))))))))).....))))..))))...................(((.....)))....))). ( -33.64) >DroSec_CAF1 33734 117 - 1 CCCUUGUGUUGGCAGCAAUUACGCUAGAAACUAUAUAUUUUGUAUAUAUAAUGUAGUUUCUGCGUCUCCCUGCUGGCAAUUCUUCUUCUUCAAAUCUUCCGAGUUUUGGUGCUGGGU (((...(((..((((.....((((.(((((((((((..............)))))))))))))))....))))..)))...........(((((.((....)).)))))....))). ( -33.44) >DroSim_CAF1 36757 117 - 1 CCCUUGUGUUGGCAGCAAUUACGCUAGAAACUAUAUAUUUUGUAUAUAUAAUGUAGUUUCUGCGUCUCCCUGCUGGCAAUUCUUCUUCUUCAAAUCUUCCGAGUUUUGGUGCUGGGU (((...(((..((((.....((((.(((((((((((..............)))))))))))))))....))))..)))...........(((((.((....)).)))))....))). ( -33.44) >DroYak_CAF1 36795 117 - 1 CCCUUGUUUUGGCAGCAAGUCCGCAAGAAACUAUAUAUUUGGUAUAUAUAAUGUAGUUUCUGCGUCUCCCUGCUGGCAAUUCUUCUUCUUCAAAUCUUCCGAGUUUUGGUGCUGAGU .....(..(((.(((((((..((((.((((((((((..............)))))))))))))).))...))))).)))..)(((..(.(((((.((....)).))))).)..))). ( -28.54) >consensus CCCUUGUGUUGGCAGCAAUUACGCUAGAAACUAUAUAUUUUGUAUAUAUAAUGUAGUUUCUGCGUCUCCCUGCUGGCAAUUCUUCUUCUUCAAAUCUUCCGAGUUUUGGUGCUGGGU ......(((..((((.....((((.(((((((((((..............)))))))))))))))....))))..)))....................((.(((......))).)). (-28.09 = -29.09 + 1.00)


| Location | 13,771,121 – 13,771,238 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -15.30 |
| Energy contribution | -17.40 |
| Covariance contribution | 2.10 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13771121 117 + 22407834 GCCAGCAGGGAGACGCAGAAACUACA---UUAUAUAUACAAAAUAUAUAGUUUCUAGCGGAAUUUCUGCCAACAAAAGGGGUCCAGUGGUUACUUCACUAGCACUCCCACACACCACCAA ....((((..(..(((((((((((..---.(((((.......))))))))))))).)))...)..))))........((((((.(((((.....))))).).)).)))............ ( -33.60) >DroSec_CAF1 33774 117 + 1 GCCAGCAGGGAGACGCAGAAACUACA---UUAUAUAUACAAAAUAUAUAGUUUCUAGCGUAAUUGCUGCCAACACAAGGGGUCCAGUGGUUACUUCACUAGCACUCCCACACACCACCAA (.((((((....((((((((((((..---.(((((.......))))))))))))).))))..)))))).).......((((((.(((((.....))))).).)).)))............ ( -33.90) >DroAna_CAF1 56185 93 + 1 ---AACCAGAAGA--CAGAAACUAAAAGUUUUAAUAACUAAAAAACACGGU--------------------ACACAAACAGUUUAGUUGUUCC-UAACCAGCACACUUAUAGGCGUCCA- ---........((--(.(((((.....)))))((((((((((.......(.--------------------...)......))))))))))((-((...((....))..)))).)))..- ( -11.52) >DroSim_CAF1 36797 117 + 1 GCCAGCAGGGAGACGCAGAAACUACA---UUAUAUAUACAAAAUAUAUAGUUUCUAGCGUAAUUGCUGCCAACACAAGGGGUCCAGUGGUUACUUCACUAGCACUCCCACACACCACCAA (.((((((....((((((((((((..---.(((((.......))))))))))))).))))..)))))).).......((((((.(((((.....))))).).)).)))............ ( -33.90) >DroYak_CAF1 36835 117 + 1 GCCAGCAGGGAGACGCAGAAACUACA---UUAUAUAUACCAAAUAUAUAGUUUCUUGCGGACUUGCUGCCAAAACAAGGGGUCCAGUGGUUACUUUACCAGCACGUCCAAACACCUCCAA ........((((..((((((((((..---.(((((.......)))))))))))).)))((((.((((.((.......))(((..(((....)))..))))))).))))......)))).. ( -33.40) >consensus GCCAGCAGGGAGACGCAGAAACUACA___UUAUAUAUACAAAAUAUAUAGUUUCUAGCGGAAUUGCUGCCAACACAAGGGGUCCAGUGGUUACUUCACUAGCACUCCCACACACCACCAA ....((((.((..(((((((((((.......................)))))))).)))...)).))))........((((((.(((((.....))))).).)).)))............ (-15.30 = -17.40 + 2.10)



| Location | 13,771,121 – 13,771,238 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -33.39 |
| Consensus MFE | -20.36 |
| Energy contribution | -22.74 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13771121 117 - 22407834 UUGGUGGUGUGUGGGAGUGCUAGUGAAGUAACCACUGGACCCCUUUUGUUGGCAGAAAUUCCGCUAGAAACUAUAUAUUUUGUAUAUAUAA---UGUAGUUUCUGCGUCUCCCUGCUGGC ..((.(((.(((((...((((.....)))).)))).).)))))....((..((((......(((.(((((((((((..............)---))))))))))))).....))))..)) ( -38.04) >DroSec_CAF1 33774 117 - 1 UUGGUGGUGUGUGGGAGUGCUAGUGAAGUAACCACUGGACCCCUUGUGUUGGCAGCAAUUACGCUAGAAACUAUAUAUUUUGUAUAUAUAA---UGUAGUUUCUGCGUCUCCCUGCUGGC ..((.(((.(((((...((((.....)))).)))).).)))))....((..((((.....((((.(((((((((((..............)---))))))))))))))....))))..)) ( -39.74) >DroAna_CAF1 56185 93 - 1 -UGGACGCCUAUAAGUGUGCUGGUUA-GGAACAACUAAACUGUUUGUGU--------------------ACCGUGUUUUUUAGUUAUUAAAACUUUUAGUUUCUG--UCUUCUGGUU--- -.(.((((......)))).).(((((-.(((((.......))))).)).--------------------)))........(((..(((((.....)))))..)))--..........--- ( -13.10) >DroSim_CAF1 36797 117 - 1 UUGGUGGUGUGUGGGAGUGCUAGUGAAGUAACCACUGGACCCCUUGUGUUGGCAGCAAUUACGCUAGAAACUAUAUAUUUUGUAUAUAUAA---UGUAGUUUCUGCGUCUCCCUGCUGGC ..((.(((.(((((...((((.....)))).)))).).)))))....((..((((.....((((.(((((((((((..............)---))))))))))))))....))))..)) ( -39.74) >DroYak_CAF1 36835 117 - 1 UUGGAGGUGUUUGGACGUGCUGGUAAAGUAACCACUGGACCCCUUGUUUUGGCAGCAAGUCCGCAAGAAACUAUAUAUUUGGUAUAUAUAA---UGUAGUUUCUGCGUCUCCCUGCUGGC ...((((.(((..(...((((.....))))....)..))).))))...(..((((..((..((((.((((((((((..............)---))))))))))))).))..))))..). ( -36.34) >consensus UUGGUGGUGUGUGGGAGUGCUAGUGAAGUAACCACUGGACCCCUUGUGUUGGCAGCAAUUACGCUAGAAACUAUAUAUUUUGUAUAUAUAA___UGUAGUUUCUGCGUCUCCCUGCUGGC ............(((.((.((((((.......)))))))))))....((..((((......(((.((((((((((....((((....))))...))))))))))))).....))))..)) (-20.36 = -22.74 + 2.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:15 2006