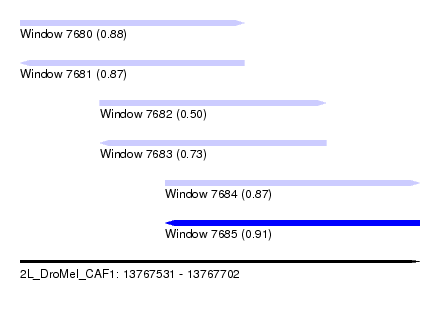
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,767,531 – 13,767,702 |
| Length | 171 |
| Max. P | 0.906126 |

| Location | 13,767,531 – 13,767,627 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.20 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -21.06 |
| Energy contribution | -20.54 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13767531 96 + 22407834 CACGGCGCAUAUGCCAGUUUAUUUAUGAUUCUG---CUCCUGGCAGCCAGAAGCAUUUAAGCACCUUAAAGUAGGCAACGCUCAGUGCUGUUGCGUCGC ..(((((((..((((................((---((.((((...)))).))))((((((...))))))...))))..((.....))...))))))). ( -28.40) >DroSec_CAF1 30165 99 + 1 CACGGCGCAUAUGGCAGUUUAUUUAUGAUUCUGCUGCUCCUGGCAGCCAGGAGCAUUUAAGCACCUUAAAGUAGGCAACGCUCGGUGCUGCUGCGUCGC ..(((((((...(((((.(((....)))..)))))((((((((...)))))))).....((((((....(((.(....)))).))))))..))))))). ( -41.30) >DroAna_CAF1 51269 97 + 1 CACGGCGCAUAUGCCAGUUUAUUUAUGGCCCUGCCAC--CAGCCCGGCGCGGGCGUUUAAGCGCCUUAAAGUACGCAAUGCGCACUGCAUUCGUGCCGC ..((((((..((((.(((.........((((((((..--......)))).))))......((((...............)))))))))))..)))))). ( -35.86) >DroSim_CAF1 33218 99 + 1 CACGGCGCAUAUGGCAGUUUAUUUAUGAUUCUGCUGCUCCUGGCAGCCAGGAGCAUUUAAGCACCUUAAAGUAGGCAACGCUCGGUGCUGCUGCGUCGC ..(((((((...(((((.(((....)))..)))))((((((((...)))))))).....((((((....(((.(....)))).))))))..))))))). ( -41.30) >DroEre_CAF1 31867 96 + 1 CACGGCGCAUAUGCCAGUUUAUUUAUGCUCCUGCU---UCCGCCGGCGAGGAGCAUUUAAGCACCUUAAAGUAGGCAACGCCCACUGCAGCUGCGUCGC ..(((((((..(((..(((((...(((((((((((---......))).)))))))).))))).......(((.(((...))).))))))..))))))). ( -38.90) >consensus CACGGCGCAUAUGCCAGUUUAUUUAUGAUUCUGCU_CUCCUGGCAGCCAGGAGCAUUUAAGCACCUUAAAGUAGGCAACGCUCAGUGCUGCUGCGUCGC ..((((((...((((.(((((...(((.(((((..............))))).))).)))))((......)).))))..((.....))....)))))). (-21.06 = -20.54 + -0.52)

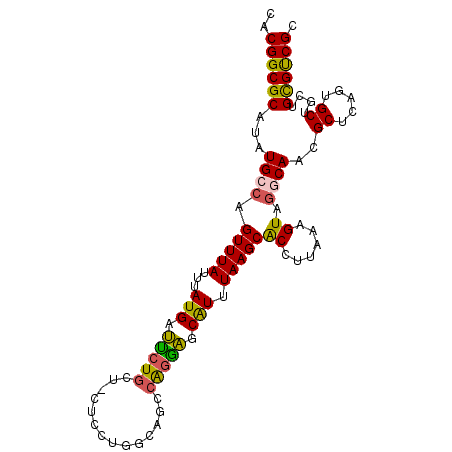
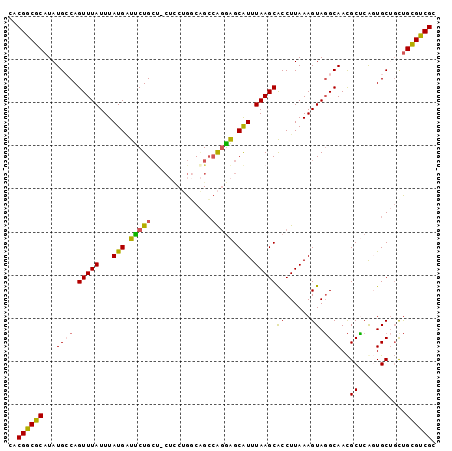
| Location | 13,767,531 – 13,767,627 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.20 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -23.70 |
| Energy contribution | -25.42 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13767531 96 - 22407834 GCGACGCAACAGCACUGAGCGUUGCCUACUUUAAGGUGCUUAAAUGCUUCUGGCUGCCAGGAG---CAGAAUCAUAAAUAAACUGGCAUAUGCGCCGUG (((.((((...((..(((((...((((......)))))))))..(((((((((...)))))))---)).................))...)))).))). ( -30.70) >DroSec_CAF1 30165 99 - 1 GCGACGCAGCAGCACCGAGCGUUGCCUACUUUAAGGUGCUUAAAUGCUCCUGGCUGCCAGGAGCAGCAGAAUCAUAAAUAAACUGCCAUAUGCGCCGUG (((((((...........))))))).........(((((.....(((((((((...)))))))))((((.............)))).....)))))... ( -37.52) >DroAna_CAF1 51269 97 - 1 GCGGCACGAAUGCAGUGCGCAUUGCGUACUUUAAGGCGCUUAAACGCCCGCGCCGGGCUG--GUGGCAGGGCCAUAAAUAAACUGGCAUAUGCGCCGUG (((((.((...(((((((((...)))))))....((((......)))).))(((((....--(((((...))))).......)))))...)).))))). ( -39.80) >DroSim_CAF1 33218 99 - 1 GCGACGCAGCAGCACCGAGCGUUGCCUACUUUAAGGUGCUUAAAUGCUCCUGGCUGCCAGGAGCAGCAGAAUCAUAAAUAAACUGCCAUAUGCGCCGUG (((((((...........))))))).........(((((.....(((((((((...)))))))))((((.............)))).....)))))... ( -37.52) >DroEre_CAF1 31867 96 - 1 GCGACGCAGCUGCAGUGGGCGUUGCCUACUUUAAGGUGCUUAAAUGCUCCUCGCCGGCGGA---AGCAGGAGCAUAAAUAAACUGGCAUAUGCGCCGUG (((.((((..((((((((((...)))))))....(((......((((((((.....((...---.))))))))))......))).)))..)))).))). ( -38.10) >consensus GCGACGCAGCAGCACUGAGCGUUGCCUACUUUAAGGUGCUUAAAUGCUCCUGGCUGCCAGGAG_AGCAGAAUCAUAAAUAAACUGGCAUAUGCGCCGUG (((((((...........))))))).........(((((.....(((((((((...))))))))).(((.............)))......)))))... (-23.70 = -25.42 + 1.72)

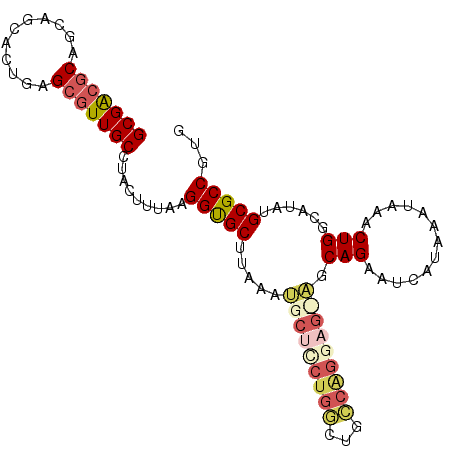
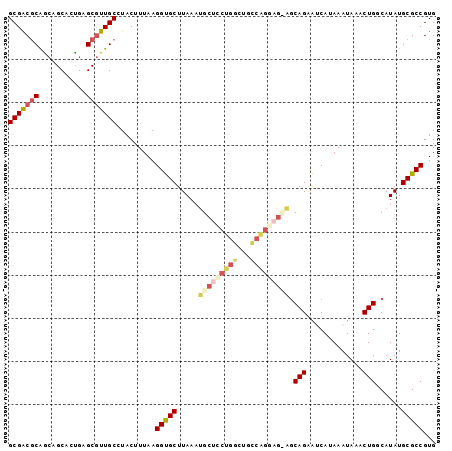
| Location | 13,767,565 – 13,767,662 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -22.02 |
| Energy contribution | -21.82 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13767565 97 + 22407834 UCCUGGCAGCCAGAAGCAUUUAAGCACCUUAAAGUAGGCAACGCUCAGUGCUGUUGCGUCGCAACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGC ..((((...))))..(((....(((((.....(((.(....))))..)))))..)))...(((((((..((((((.....)))))).))))...))) ( -27.70) >DroSec_CAF1 30202 97 + 1 UCCUGGCAGCCAGGAGCAUUUAAGCACCUUAAAGUAGGCAACGCUCGGUGCUGCUGCGUCGCAACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGC ((((((...))))))(((....((((((....(((.(....)))).))))))..)))...(((((((..((((((.....)))))).))))...))) ( -35.30) >DroAna_CAF1 51306 95 + 1 --CAGCCCGGCGCGGGCGUUUAAGCGCCUUAAAGUACGCAAUGCGCACUGCAUUCGUGCCGCAACUUCUUGCCACUUGUUGUGGCAGAUGUCGUUGC --......(((((((((((....)))))....(((.(((...))).))).....))))))(((((.(((.(((((.....))))))))....))))) ( -39.20) >DroSim_CAF1 33255 97 + 1 UCCUGGCAGCCAGGAGCAUUUAAGCACCUUAAAGUAGGCAACGCUCGGUGCUGCUGCGUCGCAACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGC ((((((...))))))(((....((((((....(((.(....)))).))))))..)))...(((((((..((((((.....)))))).))))...))) ( -35.30) >DroEre_CAF1 31902 96 + 1 -UCCGCCGGCGAGGAGCAUUUAAGCACCUUAAAGUAGGCAACGCCCACUGCAGCUGCGUCGCAACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGC -.......(((((((.(((....(((.((...(((.(((...))).)))..)).)))(((((((((..........)))))))))..)))))))))) ( -32.60) >consensus UCCUGGCAGCCAGGAGCAUUUAAGCACCUUAAAGUAGGCAACGCUCAGUGCUGCUGCGUCGCAACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGC ........((.(((((((....(((((......((.....)).....)))))..)))(((((((((..........))))))))).....)))).)) (-22.02 = -21.82 + -0.20)

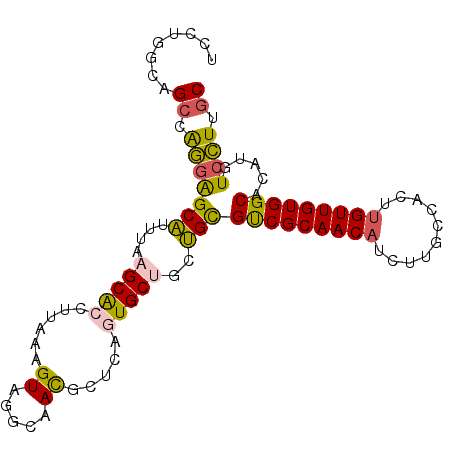
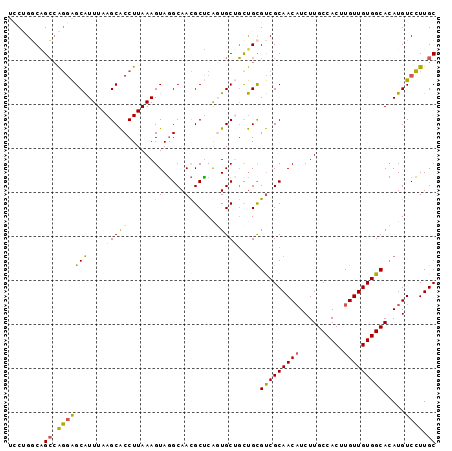
| Location | 13,767,565 – 13,767,662 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -24.68 |
| Energy contribution | -25.24 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13767565 97 - 22407834 GCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUUGCGACGCAACAGCACUGAGCGUUGCCUACUUUAAGGUGCUUAAAUGCUUCUGGCUGCCAGGA (((..(((((.((((((.....))))))..)))))(((((((..((....)).)))))))....((((((...)))))))))((((((...)))))) ( -31.20) >DroSec_CAF1 30202 97 - 1 GCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUUGCGACGCAGCAGCACCGAGCGUUGCCUACUUUAAGGUGCUUAAAUGCUCCUGGCUGCCAGGA (((.((.....((((((.....))))))...((((((......))))))))((((...((((......))))))))...)))((((((...)))))) ( -35.90) >DroAna_CAF1 51306 95 - 1 GCAACGACAUCUGCCACAACAAGUGGCAAGAAGUUGCGGCACGAAUGCAGUGCGCAUUGCGUACUUUAAGGCGCUUAAACGCCCGCGCCGGGCUG-- ((..((((.((((((((.....))))).))).))))((((.((.....(((((((...)))))))....((((......)))))).)))).))..-- ( -39.90) >DroSim_CAF1 33255 97 - 1 GCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUUGCGACGCAGCAGCACCGAGCGUUGCCUACUUUAAGGUGCUUAAAUGCUCCUGGCUGCCAGGA (((.((.....((((((.....))))))...((((((......))))))))((((...((((......))))))))...)))((((((...)))))) ( -35.90) >DroEre_CAF1 31902 96 - 1 GCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUUGCGACGCAGCUGCAGUGGGCGUUGCCUACUUUAAGGUGCUUAAAUGCUCCUCGCCGGCGGA- (((...((((.((((((.....))))))..)))))))..(((....(((((((((...)))))))...(((.((......)).))).))..)))..- ( -35.50) >consensus GCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUUGCGACGCAGCAGCACUGAGCGUUGCCUACUUUAAGGUGCUUAAAUGCUCCUGGCUGCCAGGA ((.(((((((.((((((.....))))))..)))))(((((((...........)))))))))......(((.((......)).))).))........ (-24.68 = -25.24 + 0.56)

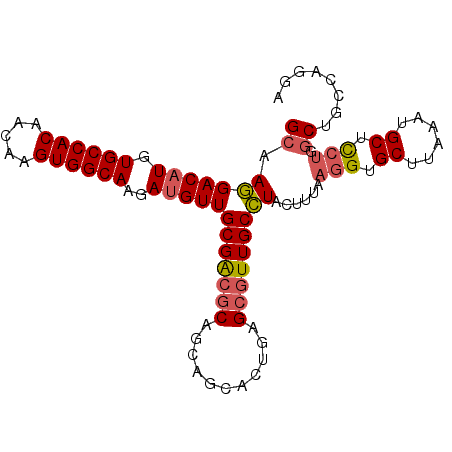
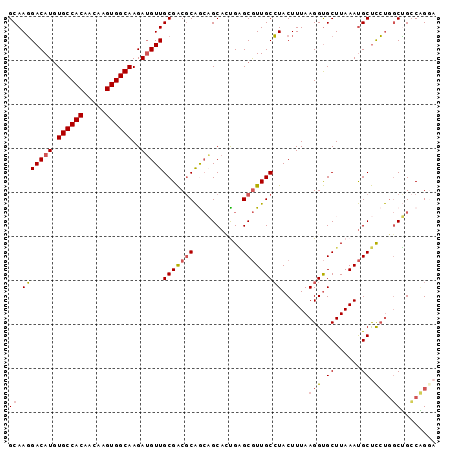
| Location | 13,767,593 – 13,767,702 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -21.21 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13767593 109 + 22407834 UUAAAGUAGGCAACGCUCAGUGCUGUUGCGUCGC-----------AACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGCUGCAACUACGAGCCGCCUCCUCGCUGCGUUUGACUUACAU .....((((((((.((.(((((..((((((..((-----------(((((..((((((.....)))))).))))...)))))))))...(((....)))..))))).))))).))))).. ( -35.10) >DroSec_CAF1 30230 96 + 1 UUAAAGUAGGCAACGCUCGGUGCUGCUGCGUCGC-----------AACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGCUGCAACCA-------------CGCUGCGUUUGACUUACAU .....(((((((((((...(((.(((.(((....-----------.((((..((((((.....)))))).))))...))).)))..))-------------)...))).))).))))).. ( -32.00) >DroAna_CAF1 51332 100 + 1 UUAAAGUACGCAAUGCGCACUGCAUUCGUGCCGC-----------AACUUCUUGCCACUUGUUGUGGCAGAUGUCGUUGCUUUU----CCAGCUCCCU---C-AUGCGU-GGGCUUACAU .....(((((.(((((.....)))))))))).((-----------(((.(((.(((((.....))))))))....)))))...(----(((((.....---.-..)).)-)))....... ( -32.90) >DroSim_CAF1 33283 96 + 1 UUAAAGUAGGCAACGCUCGGUGCUGCUGCGUCGC-----------AACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGCUGCAACCA-------------CGCUGCGUUUGACUUACAU .....(((((((((((...(((.(((.(((....-----------.((((..((((((.....)))))).))))...))).)))..))-------------)...))).))).))))).. ( -32.00) >DroEre_CAF1 31929 109 + 1 UUAAAGUAGGCAACGCCCACUGCAGCUGCGUCGC-----------AACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGCUCCAACCACGAGCCGCCUCCUCGCUGCGUUUGACUUACUU ...((((((((((.....((.(((((...(..((-----------.((((..((((((.....)))))).))))....((((.......)))).))..)...)))))))))).))))))) ( -32.10) >DroYak_CAF1 33248 120 + 1 UUAAAGUAGGCAACGCUCACUGCAGUUGCGUCGCAAGUUGGUCUCAACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGCUCCAACCACGAGCCGCCCCCUCGCUGCGUUUGACUUACAU .....(((((((((((.....((((..((((((...(((((...((((((..((((((.....)))))).))))...))..)))))..)))..)))...)).)).))).))).))))).. ( -34.80) >consensus UUAAAGUAGGCAACGCUCACUGCAGCUGCGUCGC___________AACAUCUUGCCACUUGUUGUGGCACAUGUCCUUGCUGCAACCACGAGCCGCCU___CGCUGCGUUUGACUUACAU .....(((((((((((.....((....)).................((((..((((((.....)))))).))))...............................)))).)).))))).. (-21.21 = -21.43 + 0.22)

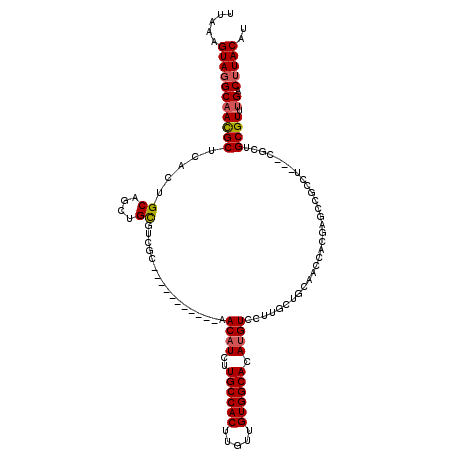
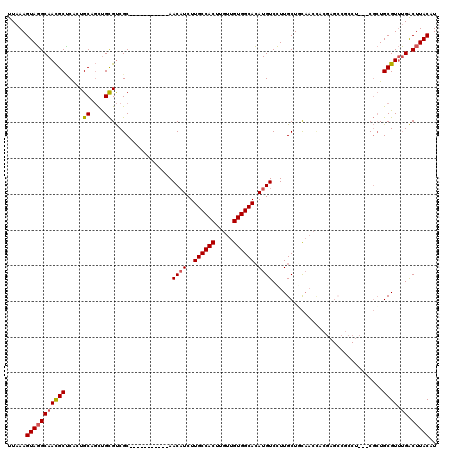
| Location | 13,767,593 – 13,767,702 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -22.87 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13767593 109 - 22407834 AUGUAAGUCAAACGCAGCGAGGAGGCGGCUCGUAGUUGCAGCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUU-----------GCGACGCAACAGCACUGAGCGUUGCCUACUUUAA ..(((.(((....(((((.(.(((....))).).))))).(((...((((.((((((.....))))))..)))))-----------)))))(((((.((.....))))))).)))..... ( -38.10) >DroSec_CAF1 30230 96 - 1 AUGUAAGUCAAACGCAGCG-------------UGGUUGCAGCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUU-----------GCGACGCAGCAGCACCGAGCGUUGCCUACUUUAA .............((((((-------------(.(((((.((..((((((.((((((.....))))))..)))))-----------.)...)).))))).....)))))))......... ( -32.90) >DroAna_CAF1 51332 100 - 1 AUGUAAGCCC-ACGCAU-G---AGGGAGCUGG----AAAAGCAACGACAUCUGCCACAACAAGUGGCAAGAAGUU-----------GCGGCACGAAUGCAGUGCGCAUUGCGUACUUUAA .(((((((((-.(....-.---.))).)))..----....((..((((.((((((((.....))))).))).)))-----------)..)).....))))((((((...))))))..... ( -34.00) >DroSim_CAF1 33283 96 - 1 AUGUAAGUCAAACGCAGCG-------------UGGUUGCAGCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUU-----------GCGACGCAGCAGCACCGAGCGUUGCCUACUUUAA .............((((((-------------(.(((((.((..((((((.((((((.....))))))..)))))-----------.)...)).))))).....)))))))......... ( -32.90) >DroEre_CAF1 31929 109 - 1 AAGUAAGUCAAACGCAGCGAGGAGGCGGCUCGUGGUUGGAGCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUU-----------GCGACGCAGCUGCAGUGGGCGUUGCCUACUUUAA .............(((((......(((((((.......))))...(((((.((((((.....))))))..)))))-----------....))).)))))(((((((...))))))).... ( -41.10) >DroYak_CAF1 33248 120 - 1 AUGUAAGUCAAACGCAGCGAGGGGGCGGCUCGUGGUUGGAGCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUUGAGACCAACUUGCGACGCAACUGCAGUGAGCGUUGCCUACUUUAA ..(((.((..(((((.(((((..(((.((((.......))))...(((((.((((((.....))))))..)))))..).))..)))))...((....)).....))))))).)))..... ( -41.30) >consensus AUGUAAGUCAAACGCAGCG___AGGCGGCUCGUGGUUGCAGCAAGGACAUGUGCCACAACAAGUGGCAAGAUGUU___________GCGACGCAACAGCACUGAGCGUUGCCUACUUUAA ..(((.(((.....((((...(((....)))...)))).......(((((.((((((.....))))))..))))).............)))(((((.((.....))))))).)))..... (-22.87 = -23.58 + 0.71)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:08 2006