
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,766,681 – 13,766,985 |
| Length | 304 |
| Max. P | 0.993882 |

| Location | 13,766,681 – 13,766,800 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -25.45 |
| Energy contribution | -26.85 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13766681 119 + 22407834 UUACAAUAUUGAUUCAUUCAUCUGAGUAAUGCCUUGGCUUUUUUAAGAGAGCGCA-AGCCACUUUUCCAAACGCUCUCUUAAGAGCUGGAAGUUUCCAGUGCUAGCCAAUGAGAGCCAAU .......((((..((((((....))))......((((((.(((((((((((((..-...............)))))))))))))((((((....))))))...))))))...))..)))) ( -35.43) >DroSec_CAF1 29312 119 + 1 UUACAGUAUUAAUUCAUUCAUCUGAGUAGUGCUUUGGCUUUUUUAAGAGAGCGCA-AGCCACCUUUCCAGCGGCUCUCUUAAUGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAU ....(((((((.((((......)))))))))))(((((((((((((((((((((.-.............)).))))))))))((((((((....))))).))).......))))))))). ( -40.94) >DroSim_CAF1 32366 119 + 1 UUACAGUAUUAAUUCAUUCAUCUGAGUAGUGCUUUGGCUUUUUUAAGAGAGCGCA-AGCCACCUUUCCAGCGGCUCUCUUAAUGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAU ....(((((((.((((......)))))))))))(((((((((((((((((((((.-.............)).))))))))))((((((((....))))).))).......))))))))). ( -40.94) >DroEre_CAF1 31081 120 + 1 UUACAAUAUUAUUUCAUUGUUUUCAAGAGUGCUUUAGAUAUUUUAAGAGAGCGCAAAGCCACUUUCCCAACAGCUCUCUUAAAGCCUGGAAAUUUCCAGUGCGAGCCAAUGAGAGCCGAU ..................(((((((...(.((((.......(((((((((((....................)))))))))))(((((((....))))).)))))))..))))))).... ( -32.25) >DroYak_CAF1 32327 120 + 1 UUGUAAUAUGAAUUCAUUCACUUGACUAGUGGCUUGUGUAUUUUAAGAGAGCGCAGAACCACUUUCCCAACAGCUCUCUUAAGGCCUGGAAAUUUCCAGAGCGAGCCAAUGAGAGCCAAU ........((((....)))).....((..((((((((...((((((((((((...(((.....)))......)))))))))))).(((((....))))).))))))))...))....... ( -34.70) >consensus UUACAAUAUUAAUUCAUUCAUCUGAGUAGUGCUUUGGCUUUUUUAAGAGAGCGCA_AGCCACUUUUCCAACAGCUCUCUUAAGGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAU ...........(((((......)))))......(((((((((((((((((((....................)))))))))).(((((((....))))).))........))))))))). (-25.45 = -26.85 + 1.40)

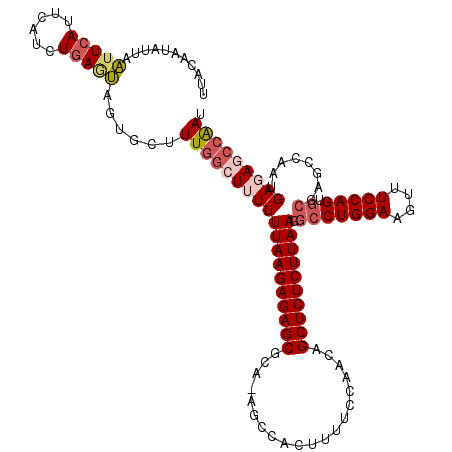

| Location | 13,766,681 – 13,766,800 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.36 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13766681 119 - 22407834 AUUGGCUCUCAUUGGCUAGCACUGGAAACUUCCAGCUCUUAAGAGAGCGUUUGGAAAAGUGGCU-UGCGCUCUCUUAAAAAAGCCAAGGCAUUACUCAGAUGAAUGAAUCAAUAUUGUAA ((((..((...((((((....(((((....)))))...((((((((((((..((........))-.))))))))))))...))))))..((((.....))))...))..))))....... ( -36.60) >DroSec_CAF1 29312 119 - 1 AUUGGCUCUCAUUGGCUCGCACUGGAAACUUCCAGGCAUUAAGAGAGCCGCUGGAAAGGUGGCU-UGCGCUCUCUUAAAAAAGCCAAAGCACUACUCAGAUGAAUGAAUUAAUACUGUAA ....(((....((((((.((.(((((....))))))).((((((((((.((.((........))-.))))))))))))...))))))))).....(((......)))............. ( -36.70) >DroSim_CAF1 32366 119 - 1 AUUGGCUCUCAUUGGCUCGCACUGGAAACUUCCAGGCAUUAAGAGAGCCGCUGGAAAGGUGGCU-UGCGCUCUCUUAAAAAAGCCAAAGCACUACUCAGAUGAAUGAAUUAAUACUGUAA ....(((....((((((.((.(((((....))))))).((((((((((.((.((........))-.))))))))))))...))))))))).....(((......)))............. ( -36.70) >DroEre_CAF1 31081 120 - 1 AUCGGCUCUCAUUGGCUCGCACUGGAAAUUUCCAGGCUUUAAGAGAGCUGUUGGGAAAGUGGCUUUGCGCUCUCUUAAAAUAUCUAAAGCACUCUUGAAAACAAUGAAAUAAUAUUGUAA ........((((((....((.(((((....)))))))((((((((.(((.((((((.((((......)))).)))))).........))).))))))))..))))))............. ( -33.90) >DroYak_CAF1 32327 120 - 1 AUUGGCUCUCAUUGGCUCGCUCUGGAAAUUUCCAGGCCUUAAGAGAGCUGUUGGGAAAGUGGUUCUGCGCUCUCUUAAAAUACACAAGCCACUAGUCAAGUGAAUGAAUUCAUAUUACAA .((((((.....(((((.((.(((((....))))))).((((((((((.((..((((.....))))))))))))))))........)))))..))))))(((((((....))).)))).. ( -38.00) >consensus AUUGGCUCUCAUUGGCUCGCACUGGAAACUUCCAGGCAUUAAGAGAGCCGUUGGAAAAGUGGCU_UGCGCUCUCUUAAAAAAGCCAAAGCACUACUCAGAUGAAUGAAUUAAUAUUGUAA ....(((....((((((.((.(((((....))))))).((((((((((..((....))((......))))))))))))...))))))))).............................. (-26.40 = -26.36 + -0.04)

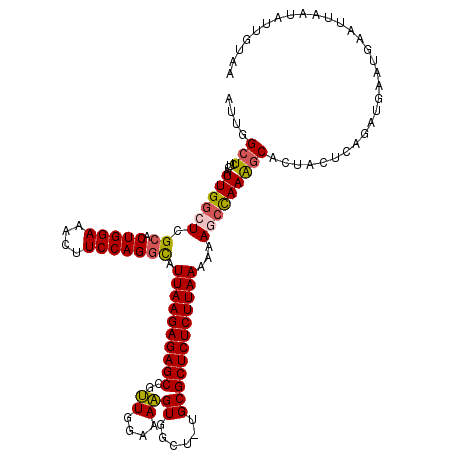

| Location | 13,766,721 – 13,766,840 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -26.69 |
| Energy contribution | -27.05 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13766721 119 + 22407834 UUUUAAGAGAGCGCA-AGCCACUUUUCCAAACGCUCUCUUAAGAGCUGGAAGUUUCCAGUGCUAGCCAAUGAGAGCCAAUUCCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGC (((((((((((((..-...............)))))))))))))((((((....))))))....(..((((((..((......)).))))))..).........((.(((....))))). ( -30.03) >DroSec_CAF1 29352 119 + 1 UUUUAAGAGAGCGCA-AGCCACCUUUCCAGCGGCUCUCUUAAUGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGC ........(.((...-.)))...........(((((((....((((((((....))))).))).......)))))))...((((((......))))))......((.(((....))))). ( -33.40) >DroSim_CAF1 32406 119 + 1 UUUUAAGAGAGCGCA-AGCCACCUUUCCAGCGGCUCUCUUAAUGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGC ........(.((...-.)))...........(((((((....((((((((....))))).))).......)))))))...((((((......))))))......((.(((....))))). ( -33.40) >DroEre_CAF1 31121 120 + 1 UUUUAAGAGAGCGCAAAGCCACUUUCCCAACAGCUCUCUUAAAGCCUGGAAAUUUCCAGUGCGAGCCAAUGAGAGCCGAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGC .(((((((((((....................)))))))))))(((((((....))))).))(((..((((((..(((....))).))))))))).........((.(((....))))). ( -33.55) >DroYak_CAF1 32367 120 + 1 UUUUAAGAGAGCGCAGAACCACUUUCCCAACAGCUCUCUUAAGGCCUGGAAAUUUCCAGAGCGAGCCAAUGAGAGCCAAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGC ..........(((((((...............((((((....((((((((....))))).....)))...))))))....((((((......))))))...............))))))) ( -33.20) >consensus UUUUAAGAGAGCGCA_AGCCACUUUUCCAACAGCUCUCUUAAGGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGC ((((((((((((....................)))))))))))).(((((....)))))..((((..((((((..((......)).))))))))))........((.(((....))))). (-26.69 = -27.05 + 0.36)

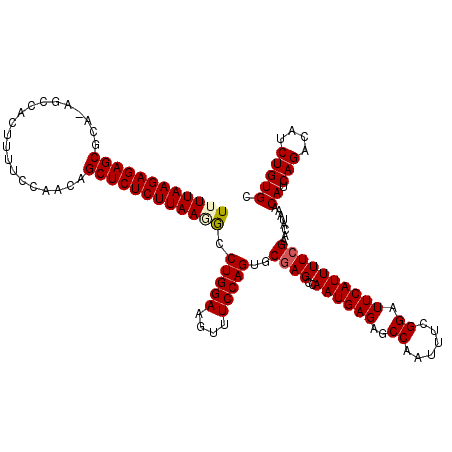
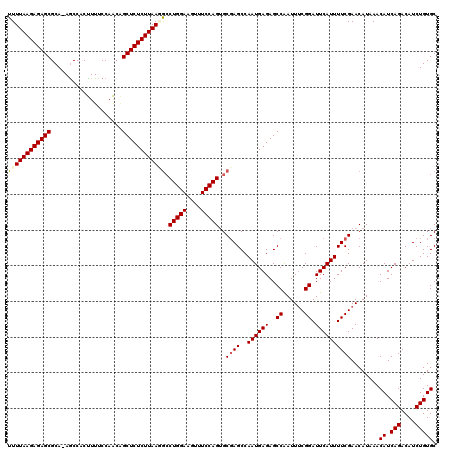
| Location | 13,766,721 – 13,766,840 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -30.20 |
| Energy contribution | -29.56 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13766721 119 - 22407834 GCACAGAUGUCUGAUGUUUAUGUUCGAAAAUGAAUCCGGAAUUGGCUCUCAUUGGCUAGCACUGGAAACUUCCAGCUCUUAAGAGAGCGUUUGGAAAAGUGGCU-UGCGCUCUCUUAAAA ((.......((((..((((((........)))))).))))..(((((......))))))).(((((....)))))...((((((((((((..((........))-.)))))))))))).. ( -34.50) >DroSec_CAF1 29352 119 - 1 GCACAGAUGUCUGAUGUUUAUGUUCGAAAAUGAAUCCGAAAUUGGCUCUCAUUGGCUCGCACUGGAAACUUCCAGGCAUUAAGAGAGCCGCUGGAAAGGUGGCU-UGCGCUCUCUUAAAA ((...((((...((.(((....((((..........))))...))))).)))).))..((.(((((....))))))).((((((((((.((.((........))-.)))))))))))).. ( -35.10) >DroSim_CAF1 32406 119 - 1 GCACAGAUGUCUGAUGUUUAUGUUCGAAAAUGAAUCCGAAAUUGGCUCUCAUUGGCUCGCACUGGAAACUUCCAGGCAUUAAGAGAGCCGCUGGAAAGGUGGCU-UGCGCUCUCUUAAAA ((...((((...((.(((....((((..........))))...))))).)))).))..((.(((((....))))))).((((((((((.((.((........))-.)))))))))))).. ( -35.10) >DroEre_CAF1 31121 120 - 1 GCACAGAUGUCUGAUGUUUAUGUUCGAAAAUGAAUCCGAAAUCGGCUCUCAUUGGCUCGCACUGGAAAUUUCCAGGCUUUAAGAGAGCUGUUGGGAAAGUGGCUUUGCGCUCUCUUAAAA ((((((....))).)))........(..(((((..(((....)))...)))))..)..((.(((((....)))))))(((((((((((.((..((.......))..))))))))))))). ( -36.40) >DroYak_CAF1 32367 120 - 1 GCACAGAUGUCUGAUGUUUAUGUUCGAAAAUGAAUCCGAAAUUGGCUCUCAUUGGCUCGCUCUGGAAAUUUCCAGGCCUUAAGAGAGCUGUUGGGAAAGUGGUUCUGCGCUCUCUUAAAA ((.((((...((....(((((........)))))(((.((...(((((((...(((.....(((((....))))))))....))))))).)).))).))....)))).)).......... ( -33.00) >consensus GCACAGAUGUCUGAUGUUUAUGUUCGAAAAUGAAUCCGAAAUUGGCUCUCAUUGGCUCGCACUGGAAACUUCCAGGCAUUAAGAGAGCCGUUGGAAAAGUGGCU_UGCGCUCUCUUAAAA ((.(((....)))............(..(((((..(((....)))...)))))..)..)).(((((....)))))...((((((((((..((....))((......)))))))))))).. (-30.20 = -29.56 + -0.64)

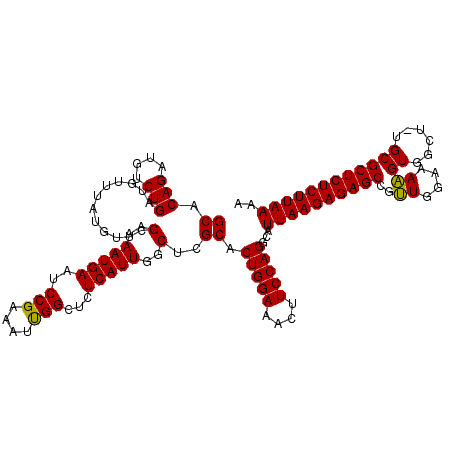

| Location | 13,766,760 – 13,766,880 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -25.96 |
| Energy contribution | -26.96 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13766760 120 + 22407834 AAGAGCUGGAAGUUUCCAGUGCUAGCCAAUGAGAGCCAAUUCCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACCAACACCAACAAAGCUCAUGGGGCCAAAAACAUGGCCA ..((((((((....))).((((..(..((((((..((......)).))))))..)............(((....))).))))............)))))....(((((......))))). ( -29.40) >DroSec_CAF1 29391 120 + 1 AAUGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCUCAUGGGGCCAAAAACAUGGCCA ..((((((((....))))).)))..(((.((((.......((((((......))))))......((.(((....))))).................)))))))(((((......))))). ( -32.20) >DroSim_CAF1 32445 120 + 1 AAUGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCUCAUGGGGCCAAAAACAUGUCCA ...(((((((....))))).))(.(((.(((((.......((((((......))))))......((.(((....))))).................)))))..))))............. ( -27.10) >DroEre_CAF1 31161 120 + 1 AAAGCCUGGAAAUUUCCAGUGCGAGCCAAUGAGAGCCGAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCUCUUGGGGCCAAAAACAUGGCCA ...(((((((....))))).))...(((...(((((....((((((......))))))......((.(((....)))))................))))))))(((((......))))). ( -33.70) >DroYak_CAF1 32407 120 + 1 AAGGCCUGGAAAUUUCCAGAGCGAGCCAAUGAGAGCCAAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCCCAUGGGGCCAAAAACAUGGCCA ...(((((((....))))).))...(((.((.(.((....((((((......))))))......((.(((....)))))................))))))))(((((......))))). ( -29.50) >consensus AAGGCCUGGAAGUUUCCAGUGCGAGCCAAUGAGAGCCAAUUUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCUCAUGGGGCCAAAAACAUGGCCA ...(((((((....))))).))...(((....((((....((((((......))))))......((.(((....)))))................)))).)))(((((......))))). (-25.96 = -26.96 + 1.00)



| Location | 13,766,800 – 13,766,918 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 97.63 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13766800 118 + 22407834 UCCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACCAACACCAACAAAGCUCAUGGGGCCAAAAACAUGGCCAAAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGACGCA ..((((......)))).............(.(.((((((((((....(((..........)))(((((......)))))....))))).......((.........)).))))).)). ( -26.20) >DroSec_CAF1 29431 118 + 1 UUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCUCAUGGGGCCAAAAACAUGGCCAAAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGACGCA ((((((......))))))...........(.(.(((((((((.....(((..........)))(((((......))))).....)))).......((.........)).))))).)). ( -24.60) >DroSim_CAF1 32485 118 + 1 UUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCUCAUGGGGCCAAAAACAUGUCCAAAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGACGCA ((((((......)))))).................(((((((.............(((.....))).......((((((....)).)))).....)))))))................ ( -19.20) >DroEre_CAF1 31201 118 + 1 UUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCUCUUGGGGCCAAAAACAUGGCCAAAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGAUGCA ((((((......))))))...........(.(((((((....................(((..(((((......)))))..)))((((((.....))))))........)))))))). ( -30.60) >DroYak_CAF1 32447 118 + 1 UUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCCCAUGGGGCCAAAAACAUGGCCAAAAGGUGCAUCCAGAGUGCAUAAAUAACAACAGACGCA ((((((......))))))...........(...(((((((((.....(((..........)))(((((......))))).....)))))..))))...)................... ( -25.50) >consensus UUCGGAUUCAUUUUCGAACAUAAACAUCAGACAUCUGUGCACAAACACCAACAAAGCUCAUGGGGCCAAAAACAUGGCCAAAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGACGCA ((((((......))))))...........(.(.(((((((((.....(((..........)))(((((......))))).....)))).......((.........)).))))).)). (-22.02 = -22.42 + 0.40)



| Location | 13,766,880 – 13,766,985 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13766880 105 + 22407834 AAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGACGCAAGAUGGACUGGCAGGCAGGUGACGAAAGUUGCAAUAUUGUUUCCACGAUGUUGAGGGGGGGGGGGGG ....((((((.....))))))......(((((..(....).((((..(((((....(..((....))..)....)))))))))...))))).............. ( -26.60) >DroSec_CAF1 29511 105 + 1 AAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGACGCAAGAUGGACUGGCAGGCAGGUGACGAAAGUUGCAAUAUUGUUUCCACGAAGGGGAGCGGUGGGGGGGG ....((((((.....))))))......(..((..(((....((((..(((((....(..((....))..)....)))))))))(....)...))).))..).... ( -27.80) >DroSim_CAF1 32565 105 + 1 AAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGACGCAAGAUGGACUGGCAGGCAGGUGACGAAAGUUGCAAUAUUGUUUCCACGAAGGGGAGCGGUGGGGGGGG ....((((((.....))))))......(..((..(((....((((..(((((....(..((....))..)....)))))))))(....)...))).))..).... ( -27.80) >DroEre_CAF1 31281 92 + 1 AAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGAUGCAAGAUGGACUGGCAGGCAGGUGACGAAAGUUGCAAUAUUGU-UCCACGAAG------------GGAGG ...(((.((((.....(((((............))))).)))).))).((((....(..((....))..)....))))-(((......------------))).. ( -22.70) >DroYak_CAF1 32527 92 + 1 AAAGGUGCAUCCAGAGUGCAUAAAUAACAACAGACGCAGGAUGGACUGGCAGGCAGGUGACGAAAGUUGCAAUAUUGU-UCCAAGAAG------------GGGGG ...(((.(((((....(((................)))))))).))).((((....(..((....))..)....))))-(((......------------))).. ( -21.99) >consensus AAAGGUGCAUCCAAAGUGCAUAAAUAACAACAGACGCAAGAUGGACUGGCAGGCAGGUGACGAAAGUUGCAAUAUUGUUUCCACGAAG__GAG_GG_GGGGGGGG ....((((((.....))))))....................((((...((((....(..((....))..)....)))).))))...................... (-20.32 = -20.32 + 0.00)

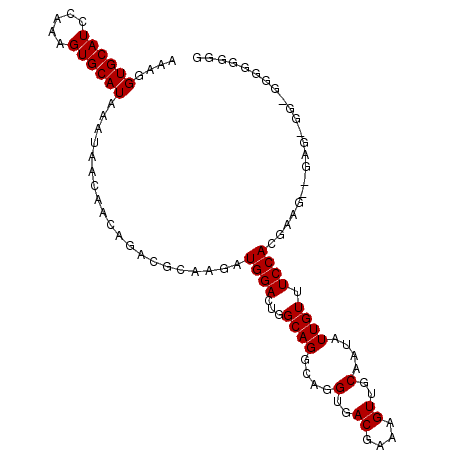

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:29:03 2006