| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,766,093 – 13,766,212 |
| Length | 119 |
| Max. P | 0.506729 |

| Location | 13,766,093 – 13,766,212 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -13.89 |
| Energy contribution | -15.95 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.46 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
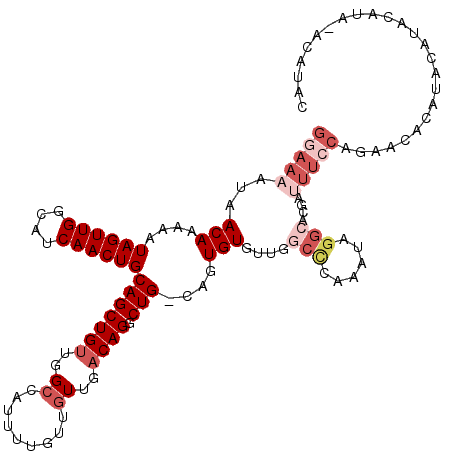
>2L_DroMel_CAF1 13766093 119 - 22407834 GGAAAAUAACAAAAAUAGUUGGCAUCAACUGCAGCUGUUGGCCAUUUUGUUGUUGACAGGCUG-CAGUGUGUUGGCCCAAAUAGGCACGAUUUCCAGAACACAUACAUACAUACAUAUAC (((((............((..((((..(((((((((((..((.........))..))).))))-))))))))..))((.....)).....)))))......................... ( -33.50) >DroSec_CAF1 28757 119 - 1 GGAAAAUAACAAAAAUAGUUGGCAUCAACUGCAGCUGUUGGCCGUUUUGUUGUUGACAGGCUG-CACUGUGUUGGCCCAAAUAGGCACGAUUUCCAGAACACAUACAUACAUAUACACAC (((((.(((((.....(((((....)))))((((((((..((.........))..))).))))-)....)))))(((......)))....)))))......................... ( -30.30) >DroAna_CAF1 50163 110 - 1 AAAAAAUAACAAAAAUAGUUGGCAUCAACUGCAGCUGGCGGAAACUUUGUUGUUGACAGACUGGCAUUGUUUU-CCCCACAUAU---------ACUAAACUCUUAUAUGGGCACACAUAA ...............((((((....))))))..(((((.((((((..(((((((....))).))))..)))))-).)).(((((---------(.........)))))))))........ ( -22.70) >DroSim_CAF1 31813 118 - 1 GGAAAAUAACAAAAAUAGUUGGCAUCAACUGCAGCUGUUGGCCGUUUUGUUGUUGACAGGCUG-CACUGUGUUG-CCCAAAUAGGCAGCGAUUCCUGAACACAUACAUACAUAUACACAC ((((....(((.....(((((....)))))((((((((..((.........))..))).))))-)..)))((((-((......))))))..))))......................... ( -32.00) >DroEre_CAF1 30534 102 - 1 GGAAAAUAACAAAAAUAGUUGGCAUCAACUGCAGCUGUUGGCCAUUUUGUUGUUGACAGGCUG-CAGUGU---GGCUCAAAUAGGCACGAUUUCCCGCACAU--------------AUAC (((((.((((.......))))...((.(((((((((((..((.........))..))).))))-))))((---(.((.....)).)))))))))).......--------------.... ( -29.70) >DroYak_CAF1 31704 102 - 1 GAAAAAUAACAAAAAUAGUUGGCAUCAACUGCAGCUGUUGGCCAUUUUGCUGUUGACAGGCUG-CAGUGU---GGCCCAAAUAGGCACGAUUUCCAGCACAU--------------AUAC .................(((((.(((.(((((((((((..((.........))..))).))))-))))((---(.((......))))))))..)))))....--------------.... ( -33.10) >consensus GGAAAAUAACAAAAAUAGUUGGCAUCAACUGCAGCUGUUGGCCAUUUUGUUGUUGACAGGCUG_CAGUGUGUUGGCCCAAAUAGGCACGAUUUCCAGAACACAUACAUACAUA_ACAUAC (((((...(((....((((((....))))))(((((((..((.........))..)))).)))....)))....(((......)))....)))))......................... (-13.89 = -15.95 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:57 2006