
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,761,410 – 13,761,502 |
| Length | 92 |
| Max. P | 0.855985 |

| Location | 13,761,410 – 13,761,502 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -14.14 |
| Energy contribution | -15.86 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
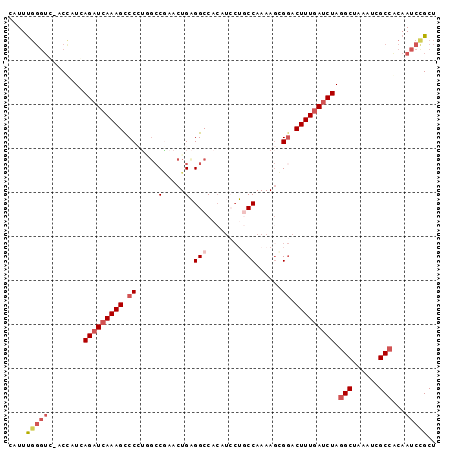
>2L_DroMel_CAF1 13761410 92 + 22407834 CAUUUC-----ACCAUCAGAUCAAAGUCCCUGGCCAAACUGAGGCCACAUCCUGCCAAAAGGGGACUUUGAUCUAGGCUAAAUCGCCACAAUCCGCU ......-----......((((((((((((((((((.......)))))....((......))))))))))))))).(((......))).......... ( -34.40) >DroSec_CAF1 25004 97 + 1 CAUUUGGGUGAACUUUCAGAUCAAAGCCCCUCGCCGAACUGAGGCCACAUCCUGCCAAAAGCGGACUUUGAUCUAGGCUAAAUCGCCACAAUCCGCU .....((((........(((((((((..(((((......))))).......((((.....)))).))))))))).(((......)))...))))... ( -27.60) >DroSim_CAF1 28060 97 + 1 CAUUUGGGUGAACUUUCAGAUCAAAGCCCCUGUCCGAACUGAGGCCACAUCCUGCCAAAAGCGGACUUUGAUCUAGGCUAAAUCGCCACAAUCCGCU .....((((........((((((((......(((((......(((........))).....))))))))))))).(((......)))...))))... ( -26.10) >DroEre_CAF1 26730 96 + 1 AAUUCGGGUC-ACCAUCAGAUCAAAGUCCAAUGGCUAACUGCGGCCACAUGCGUCCAAAAAGGGACUUUGAUCUAGGCUAAAUCGCCGCAAUCUGCU .....((...-.))...((((((((((((..(((((......)))))..((....)).....)))))))))))).(((......)))((.....)). ( -31.20) >DroYak_CAF1 28122 89 + 1 A-UUUGGGUC-ACCAUGAGAUGAAAGCCCAAAGCCGAACUGCGGCC------GUCCAAAAGCGCACUUUGAGCUAAGCUAAAUCGCCACAAUCCGCU .-(((((((.-..(((...)))...)))))))((((.....)))).------.......((((.....((.((...........)))).....)))) ( -18.00) >consensus CAUUUGGGUC_ACCAUCAGAUCAAAGCCCCUGGCCGAACUGAGGCCACAUCCUGCCAAAAGCGGACUUUGAUCUAGGCUAAAUCGCCACAAUCCGCU ....(((((........(((((((((.((.....(.....).(((........)))......)).))))))))).(((......)))...))))).. (-14.14 = -15.86 + 1.72)


| Location | 13,761,410 – 13,761,502 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.74 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
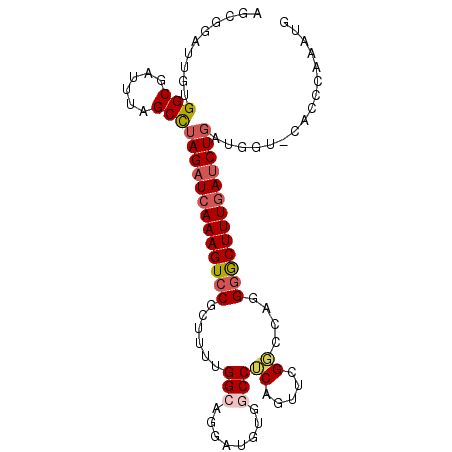
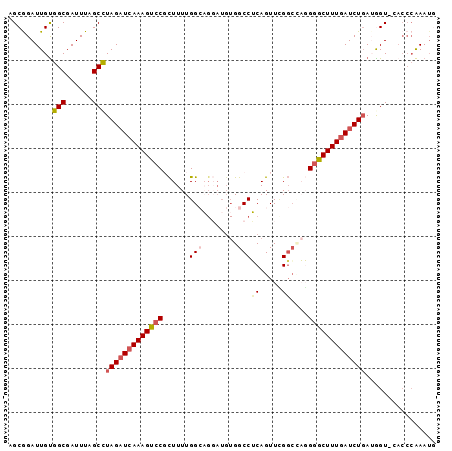
>2L_DroMel_CAF1 13761410 92 - 22407834 AGCGGAUUGUGGCGAUUUAGCCUAGAUCAAAGUCCCCUUUUGGCAGGAUGUGGCCUCAGUUUGGCCAGGGACUUUGAUCUGAUGGU-----GAAAUG .....((..((((......)))(((((((((((((((((.....)))...(((((.......))))))))))))))))))))..))-----...... ( -35.50) >DroSec_CAF1 25004 97 - 1 AGCGGAUUGUGGCGAUUUAGCCUAGAUCAAAGUCCGCUUUUGGCAGGAUGUGGCCUCAGUUCGGCGAGGGGCUUUGAUCUGAAAGUUCACCCAAAUG ...(((((..(((......)))(((((((((((((((.....))........(((.......)))...)))))))))))))..)))))......... ( -32.70) >DroSim_CAF1 28060 97 - 1 AGCGGAUUGUGGCGAUUUAGCCUAGAUCAAAGUCCGCUUUUGGCAGGAUGUGGCCUCAGUUCGGACAGGGGCUUUGAUCUGAAAGUUCACCCAAAUG ...(((((..(((......)))(((((((((((((.(((((((((((......)))..).))))).)))))))))))))))..)))))......... ( -30.20) >DroEre_CAF1 26730 96 - 1 AGCAGAUUGCGGCGAUUUAGCCUAGAUCAAAGUCCCUUUUUGGACGCAUGUGGCCGCAGUUAGCCAUUGGACUUUGAUCUGAUGGU-GACCCGAAUU ....(((((((((((((((...))))))...((((......)))).......))))))))).(((((..((......))..)))))-.......... ( -33.30) >DroYak_CAF1 28122 89 - 1 AGCGGAUUGUGGCGAUUUAGCUUAGCUCAAAGUGCGCUUUUGGAC------GGCCGCAGUUCGGCUUUGGGCUUUCAUCUCAUGGU-GACCCAAA-U ..(((((((((((..(((((...(((.(.....).))).))))).------.)))))))))))..((((((...(((((....)))-))))))))-. ( -32.40) >consensus AGCGGAUUGUGGCGAUUUAGCCUAGAUCAAAGUCCGCUUUUGGCAGGAUGUGGCCUCAGUUCGGCCAGGGGCUUUGAUCUGAUGGU_CACCCAAAUG ..........(((......)))(((((((((((((......(((........)))((.....))....)))))))))))))................ (-19.98 = -20.74 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:53 2006