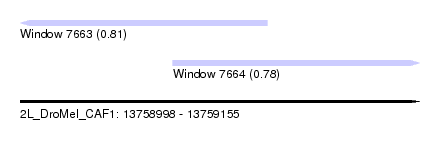
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,758,998 – 13,759,155 |
| Length | 157 |
| Max. P | 0.809985 |

| Location | 13,758,998 – 13,759,095 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.87 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13758998 97 - 22407834 GAAA---AAAGUCGAGAAGUCGGCCGACUAAUCAAAUUACCGCAAUGG---------------GAAGUUGGCAAUUGAAAGCUUAGGACUUUAGGAUGUAGCCUCAGUCUUGAGU----- ....---.....(((....)))(((((((..(((...........)))---------------..)))))))........((((((((((..(((......))).))))))))))----- ( -25.70) >DroSec_CAF1 22631 95 - 1 --AA---AAAGUCGAGAAGUCGGCCGACUAAUCAAAUUACCGCAAUGG---------------GAAGUUGGCAAUUGAAAGCUUAGGACUUUAGGAUGUAGCCUUAGUCUUGAGU----- --..---.....(((....)))(((((((..(((...........)))---------------..)))))))........((((((((((..(((......))).))))))))))----- ( -26.70) >DroAna_CAF1 43523 120 - 1 GAAAAAAUAAGUUAAAAAGUCGGCCGACUAAUCAAAUUACGGCAAUGAUGCUUGCCACAAAAGGAAAAUGGAGAUAAAAACAUUGUGGGUGUAGGGGCAAGUCUUGGUGGAAAGUUUGAG .................((((....))))..(((((((.((.(((....((((((((((.......((((..........)))).....)))...))))))).))).))...))))))). ( -22.90) >DroSim_CAF1 25660 97 - 1 GAAA---AAAGUCGAGAAGUCGGCCGACUAAUCAAAUUACCGCAAUGG---------------GAAGUUGGCAAUUGAAAGCUUAGGACUUUAGGAUGUAGUCUUAGUCUUGAGU----- ....---.....(((....)))(((((((..(((...........)))---------------..)))))))........((((((((((..(((((...)))))))))))))))----- ( -24.60) >DroEre_CAF1 24295 97 - 1 GAAA---AAAGUCGACAAGCCGGCCGACUAAUCAAAUUACCGCAAUGG---------------GAAGUUGGCAAUUGCAAGCUUAGGACUUUGGGAUGUAGCCUUAGUCUUGAGU----- ....---...........((..(((((((..(((...........)))---------------..)))))))....))..((((((((((..(((......))).))))))))))----- ( -27.60) >DroYak_CAF1 25745 99 - 1 GAAA-AAAAAGUCGAGAAGUCGGCCGACUAAUCAAAUUACCGCAAUGG---------------AGAGUUGGCAGUUGAAAGCUUAGGACUUUGGGCUGUAGCCUUAGUCUUGAGU----- ....-.......(((....)))(((((((..(((...........)))---------------..)))))))........((((((((((..((((....)))).))))))))))----- ( -30.10) >consensus GAAA___AAAGUCGAGAAGUCGGCCGACUAAUCAAAUUACCGCAAUGG_______________GAAGUUGGCAAUUGAAAGCUUAGGACUUUAGGAUGUAGCCUUAGUCUUGAGU_____ ......................(((((((....................................)))))))........((((((((((..(((......))).))))))))))..... (-16.67 = -16.87 + 0.20)


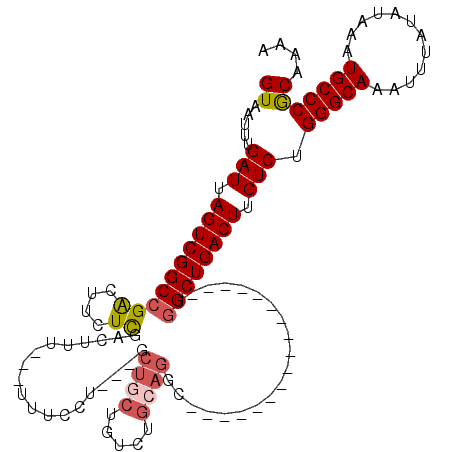
| Location | 13,759,058 – 13,759,155 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.63 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -23.63 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13759058 97 + 22407834 GUAAUUUGAUUAGUCGGCCGACUUCUCGACUUU---UUUCCU---GCUCCUGUCUGCAGGC-----------------UGGCUGACUUGUCUGGGCAAAUUUAUAUAAAUGCCCGCAAAA ((.....(((.((((((((((....))......---...(((---((........))))).-----------------.)))))))).))).(((((............))))))).... ( -31.60) >DroSec_CAF1 22691 95 + 1 GUAAUUUGAUUAGUCGGCCGACUUCUCGACUUU---UU--CU---CCUGCUGUCUGCAGGC-----------------UGGCUGACUUGUCUGGGCAAAUUUAUAUAAAUGCCCGCAAAA ((.....(((.((((((((((....))......---..--..---(((((.....))))).-----------------.)))))))).))).(((((............))))))).... ( -31.60) >DroAna_CAF1 43603 120 + 1 GUAAUUUGAUUAGUCGGCCGACUUUUUAACUUAUUUUUUCCUUCAGCUCCUGUCUGCAGGUCGACUUUCUUCUCCCUUUUGCUGACUUGUCUGGGCAAAUUUAUAUAAAUGCCCACAAAA ((((.((((..((((....))))..)))).))))........(((((.((((....))))..((......))........))))).((((..(((((............))))))))).. ( -25.30) >DroSim_CAF1 25720 97 + 1 GUAAUUUGAUUAGUCGGCCGACUUCUCGACUUU---UUUCCU---GCUGCUGUCUGCAGGC-----------------UGGCUGACUUGUCUGGGCAAAUUUAUAUAAAUGCCCGCAAAA ((.....(((.((((((((((....))......---...(((---((........))))).-----------------.)))))))).))).(((((............))))))).... ( -31.60) >DroEre_CAF1 24355 97 + 1 GUAAUUUGAUUAGUCGGCCGGCUUGUCGACUUU---UUUCGU---GCUGCUGUCUGCCUGC-----------------UGGCUGACUUGUCUGGGCAAAUUUAUAUAAAUGCCCGCAAAA ((.....(((.(((((((((((..((.(((...---......---......))).))..))-----------------))))))))).))).(((((............))))))).... ( -35.16) >DroYak_CAF1 25805 99 + 1 GUAAUUUGAUUAGUCGGCCGACUUCUCGACUUUUU-UUUCGU---GCUGAUGUCUGCAGGC-----------------UGGCUGACUUGUCUGGGCAAAUUUAUAUAAAUGCCCGCAAAA ((.....(((.(((((((((.((...(((......-..)))(---((........))))).-----------------))))))))).))).(((((............))))))).... ( -27.90) >consensus GUAAUUUGAUUAGUCGGCCGACUUCUCGACUUU___UUUCCU___GCUGCUGUCUGCAGGC_________________UGGCUGACUUGUCUGGGCAAAUUUAUAUAAAUGCCCGCAAAA ((.....(((.((((((((((....))...................((((.....))))....................)))))))).))).(((((............))))))).... (-23.63 = -24.22 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:49 2006