| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,757,699 – 13,757,806 |
| Length | 107 |
| Max. P | 0.519548 |

| Location | 13,757,699 – 13,757,806 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.29 |
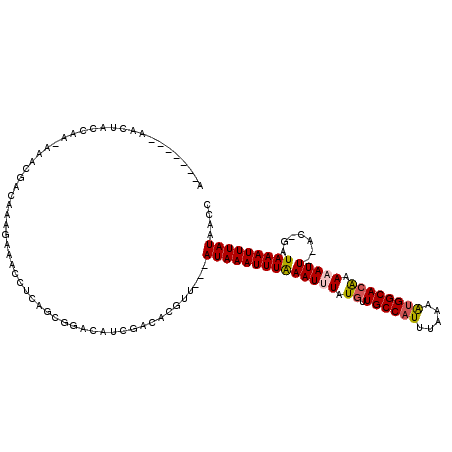
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -10.18 |
| Energy contribution | -10.43 |
| Covariance contribution | 0.25 |
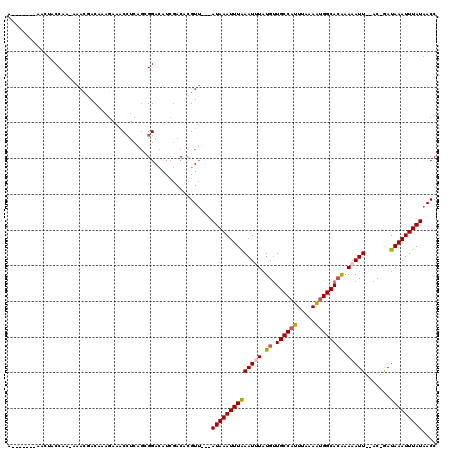
| Combinations/Pair | 1.14 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
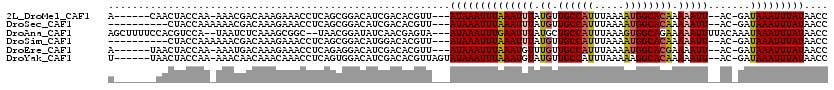
>2L_DroMel_CAF1 13757699 107 - 22407834 A------CAACUACCAA-AAACGACAAAGAAACCUCAGCGGACAUCGACACGUU---AUAAAUUUAAAUUUAUGUUGCCAUUUAAAAUGGCACAAAAAUU--AC-GAUAAAUUUAUAACC .------..........-..............((.....))..........(((---((((((((((((((.(((.(((((.....)))))))).)))))--..-..)))))))))))). ( -19.30) >DroSec_CAF1 21313 104 - 1 ----------CUACCAAAAAACGACAAAGAAACCUCAGCGGACAUCGACACGUU---AUAAAUUUAAAUUUAUGUUGCCAUUUAAAAUGGCACAAAAAUU--AC-GAUAAAUUUAUAACC ----------......................((.....))..........(((---((((((((((((((.(((.(((((.....)))))))).)))))--..-..)))))))))))). ( -19.30) >DroAna_CAF1 42051 113 - 1 AGCUUUUCCACGUCCA--UAAUCUCAAAGCGGC--UAACGGAUAUCAACGAGUA---AUAAAUUUGAAUUUAUGCUGCCAUUUAAAGUGGCAGAAAAAUUUUACAAAUAAAUUUAUAACC ..............((--(((..(((((...((--(..((........))))).---.....)))))..)))))(((((((.....)))))))..((((((.......))))))...... ( -19.10) >DroSim_CAF1 23417 104 - 1 ----------CUACCAAAAAACGACAAAGAAACCUCAGCGGACAUGGACACGUU---AUAAAUUUAAAUUUAUGUUGCCAUUUAAAAUGGCACAAAAAUU--AC-GAUAAAUUUAUAACC ----------...(((.....((.(..((....))..)))....)))....(((---((((((((((((((.(((.(((((.....)))))))).)))))--..-..)))))))))))). ( -21.50) >DroEre_CAF1 23118 107 - 1 A------UAACUACCAA-AAAUGACAAAGAAACCUCAGAGGACAUCGACACGUU---AUAAAUUUAAAUGUUUGUUGCCAUUUAAAAUGGCACGAAAAUU--AC-GAUAAAUUUAUAACC .------..........-..............((.....))..........(((---((((((((((((.(((((.(((((.....)))))))))).)))--..-..)))))))))))). ( -19.30) >DroYak_CAF1 24514 110 - 1 U------UAACUACCAA-AAACAACAAACAAACCUCAGUGGACAUCGACACGUUAGUAUAAAUUUAAAUGUAUGUUGCCAUUUAAAAAGGCACAAAAAUU--AC-GAUAAAUUUAUAACC .------..........-...................(((........))).....((((((((((..(((((((.(((.........)))))).....)--))-).))))))))))... ( -13.60) >consensus A_______AACUACCAA_AAACGACAAAGAAACCUCAGCGGACAUCGACACGUU___AUAAAUUUAAAUUUAUGUUGCCAUUUAAAAUGGCACAAAAAUU__AC_GAUAAAUUUAUAACC .........................................................((((((((((((((.((.((((((.....)))))))).))))).......))))))))).... (-10.18 = -10.43 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:45 2006