
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,756,273 – 13,756,497 |
| Length | 224 |
| Max. P | 0.997920 |

| Location | 13,756,273 – 13,756,377 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -24.15 |
| Energy contribution | -23.78 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13756273 104 + 22407834 AAGGAUGGACGGAAGCAUCCAGCUUGUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGCGCCGGCGAGAGGCAAAAAUACAAAAAAA ..(((((..(....))))))...((((((((.((((.((((..((((...))))..((((....))))....))))))))...))))))))............. ( -30.60) >DroSim_CAF1 21973 102 + 1 AAGGAUGGACGGAAGCAUCCAGCUUGUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGCGGCGGCGAGAGGAAAAAUUAAAAAA--A ..(((((..(....))))))..((((((......(((((((..((((...))))..((((....))))....)))))))))))))................--. ( -29.20) >DroEre_CAF1 21733 95 + 1 AAGGAUGGACGGAAGCAUCCUGCUUGUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGC---GGCGAGAGGCAAAAA----AU--AA .((((((..(....)))))))..((((((((...(((((((..((((...))))..((((....))))....))))---))).))))))))...----..--.. ( -33.30) >DroYak_CAF1 23058 91 + 1 AAGGAUGAACGGAAGUAUCCUGCUCGUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGC---GGCGAGAGGCAAAAA---------- .(((((..((....)))))))....((((((...(((((((..((((...))))..((((....))))....))))---))).)))))).....---------- ( -31.00) >consensus AAGGAUGGACGGAAGCAUCCAGCUUGUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGC___GGCGAGAGGCAAAAA____AA___A ..(((((..(....))))))..((((((......((((((...((((...))))..((((....)))).).)))))...))))))................... (-24.15 = -23.78 + -0.37)


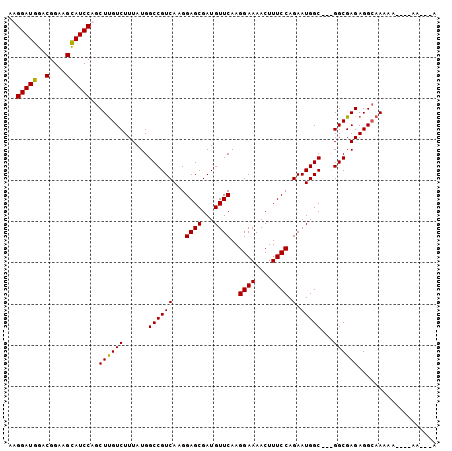
| Location | 13,756,298 – 13,756,417 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -12.27 |
| Energy contribution | -13.64 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13756298 119 + 22407834 GUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGCGCCGGCGAGAGGCAAAAAUACAAAAAAAAAAAAGAAUACGAAAAUUGGCACAUAAAAAGUUUUUUAAU ...(((((((((((((..((((...))))..((((....))))....))))(((.......))).................................))).))))))............ ( -23.80) >DroAna_CAF1 40699 93 + 1 GUCUUUAUGGCUGCCAAGUAGCU---------GCAAACUUU--------GC---GACGGAAGGCAGGAA----GU--AGAACGACAAAAAAAAUAUUGGCAUAUAAAAAGUUUUUUAAU ((((((((..(((((.....(.(---------(((.....)--------))---).)....)))))...----))--)))..)))..(((((((.((.........)).)))))))... ( -15.90) >DroSim_CAF1 21998 117 + 1 GUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGCGGCGGCGAGAGGAAAAAUUAAAAAA--AAAAAAGAAUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAU .(((((...(((((((..((((...))))..((((....))))....)))))))..(....)..............--...)))))...((((((((...........))))))))... ( -22.30) >DroEre_CAF1 21758 110 + 1 GUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGC---GGCGAGAGGCAAAAA----AU--AAAAAAAAAAUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAU ((((((...(((((((..((((...))))..((((....))))....))))---))).)))))).....----..--............((((((((...........))))))))... ( -25.20) >DroYak_CAF1 23083 99 + 1 GUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGC---GGCGAGAGGCAAAAA-----------------AUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAU ((((((...(((((((..((((...))))..((((....))))....))))---))).)))))).....-----------------...((((((((...........))))))))... ( -25.20) >consensus GUCUUUAUGGCCGUCAAGGAGCGAUGUUCAAGGAAAACUUUCCAGAAUGGC___GGCGAGAGGCAAAAA____AA__AAAAAAAGAAUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAU ...(((((((((((((..((((...))))..((((....))))....))))...(.(....).).................................))).))))))............ (-12.27 = -13.64 + 1.37)

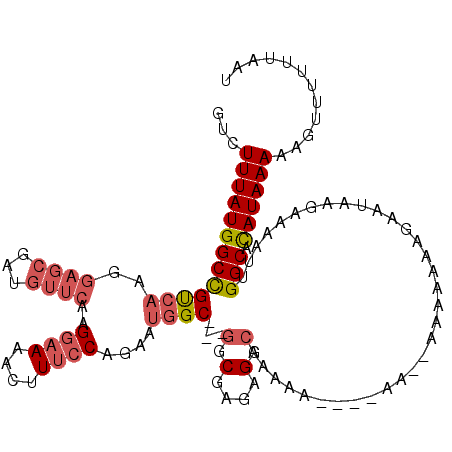
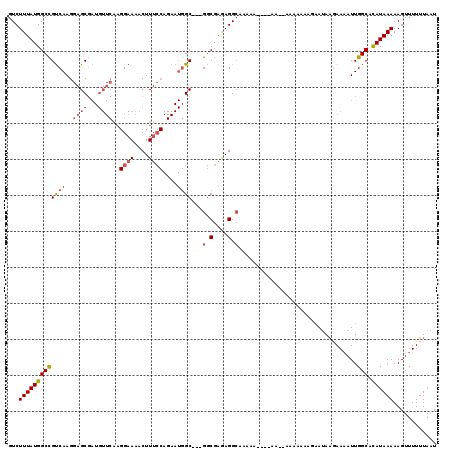
| Location | 13,756,377 – 13,756,497 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.85 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13756377 120 + 22407834 AAAAAGAAUACGAAAAUUGGCACAUAAAAAGUUUUUUAAUGAAUGGAAUUAGUGUUUGAGCUGCAGCGACUUUGCUAAAUCGUCCGAGCAUCGCCUCGGAAGUCCACUCAGUGUGCCACU .................((((((((((((....))))..(((.((((.((((((.....((....)).....))))))....((((((......))))))..)))).))))))))))).. ( -31.20) >DroSec_CAF1 19967 120 + 1 AAAAAGAAUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAUGAAUGGAAUUAGUGUUUGAGCUGCAGCGACUUUGCUAAAUCGUCCGAGCAUCGCCUCGGAAGUCCACUCAGAGUGCCACU .................((((((.(((((....))))).(((.((((.((((((.....((....)).....))))))....((((((......))))))..)))).)))..)))))).. ( -30.60) >DroAna_CAF1 40752 120 + 1 AACGACAAAAAAAAUAUUGGCAUAUAAAAAGUUUUUUAAUGAAUGGAAUUAGUGUUUGAGCUGCACCGACUUUGUUAAAUCGUCCGAGCAGCGUCUCGGAAGUCCACUCAGAGUCCUUCU ...(((..(((((((.((.........)).)))))))..(((.((((............(((((.(.(((...........))).).)))))....(....))))).)))..)))..... ( -22.50) >DroSim_CAF1 22075 120 + 1 AAAAAGAAUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAUGAAUGGAAUUAGUGUUUGAGCUGCAGCGACUUUGCUAAAUCGUCCGAGCAUCGCCUCGGAAGUCCACUCAGAGUGCCACU .................((((((.(((((....))))).(((.((((.((((((.....((....)).....))))))....((((((......))))))..)))).)))..)))))).. ( -30.60) >DroEre_CAF1 21828 120 + 1 AAAAAAAAUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAUGAAUGGAAUUAGUGUUUGAGCUGCAGCGACUUUGCUAAAUCGUCCGAGCAUCGCCUCGGAAGUCCACUCAGAGUGCCACU .................((((((.(((((....))))).(((.((((.((((((.....((....)).....))))))....((((((......))))))..)))).)))..)))))).. ( -30.60) >DroYak_CAF1 23149 113 + 1 -------AUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAUGAAUGGAAUUAGUGUUUGAGCUGCAGCGACUUUGCUAAAUCGUCCGAGCAUCGCCUCGGAAGUCCACUCAGAGUGCCACU -------..........((((((.(((((....))))).(((.((((.((((((.....((....)).....))))))....((((((......))))))..)))).)))..)))))).. ( -30.60) >consensus AAAAAGAAUAAGAAAAUUGGCACAUAAAAAGUUUUUUAAUGAAUGGAAUUAGUGUUUGAGCUGCAGCGACUUUGCUAAAUCGUCCGAGCAUCGCCUCGGAAGUCCACUCAGAGUGCCACU .................((((((.(((((....))))).(((.((((.((((((.....((....)).....))))))....((((((......))))))..)))).)))..)))))).. (-26.76 = -26.85 + 0.09)

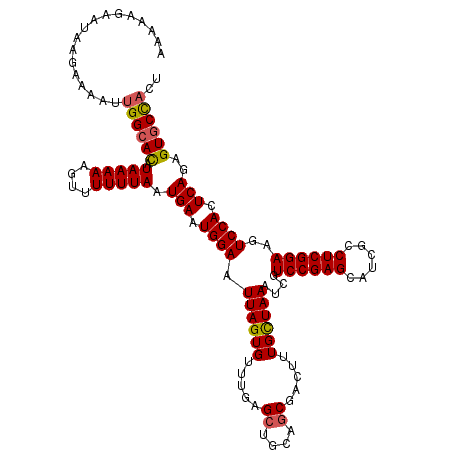
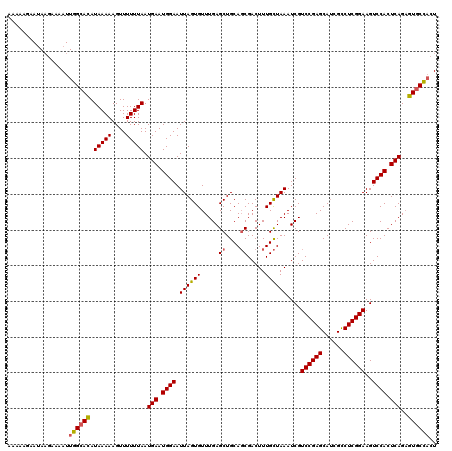
| Location | 13,756,377 – 13,756,497 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -28.52 |
| Energy contribution | -29.18 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13756377 120 - 22407834 AGUGGCACACUGAGUGGACUUCCGAGGCGAUGCUCGGACGAUUUAGCAAAGUCGCUGCAGCUCAAACACUAAUUCCAUUCAUUAAAAAACUUUUUAUGUGCCAAUUUUCGUAUUCUUUUU ..(((((((.((((((((..((((((......))))))((((((....))))))...................)))))))).(((((....))))))))))))................. ( -32.80) >DroSec_CAF1 19967 120 - 1 AGUGGCACUCUGAGUGGACUUCCGAGGCGAUGCUCGGACGAUUUAGCAAAGUCGCUGCAGCUCAAACACUAAUUCCAUUCAUUAAAAAACUUUUUAUGUGCCAAUUUUCUUAUUCUUUUU ..((((((..((((((((..((((((......))))))((((((....))))))...................)))))))).(((((....))))).))))))................. ( -31.60) >DroAna_CAF1 40752 120 - 1 AGAAGGACUCUGAGUGGACUUCCGAGACGCUGCUCGGACGAUUUAACAAAGUCGGUGCAGCUCAAACACUAAUUCCAUUCAUUAAAAAACUUUUUAUAUGCCAAUAUUUUUUUUGUCGUU ..((.(((..((((((((..((((((......))))))((((((....))))))...................))))))))..............((((....)))).......))).)) ( -25.30) >DroSim_CAF1 22075 120 - 1 AGUGGCACUCUGAGUGGACUUCCGAGGCGAUGCUCGGACGAUUUAGCAAAGUCGCUGCAGCUCAAACACUAAUUCCAUUCAUUAAAAAACUUUUUAUGUGCCAAUUUUCUUAUUCUUUUU ..((((((..((((((((..((((((......))))))((((((....))))))...................)))))))).(((((....))))).))))))................. ( -31.60) >DroEre_CAF1 21828 120 - 1 AGUGGCACUCUGAGUGGACUUCCGAGGCGAUGCUCGGACGAUUUAGCAAAGUCGCUGCAGCUCAAACACUAAUUCCAUUCAUUAAAAAACUUUUUAUGUGCCAAUUUUCUUAUUUUUUUU ..((((((..((((((((..((((((......))))))((((((....))))))...................)))))))).(((((....))))).))))))................. ( -31.60) >DroYak_CAF1 23149 113 - 1 AGUGGCACUCUGAGUGGACUUCCGAGGCGAUGCUCGGACGAUUUAGCAAAGUCGCUGCAGCUCAAACACUAAUUCCAUUCAUUAAAAAACUUUUUAUGUGCCAAUUUUCUUAU------- ..((((((..((((((((..((((((......))))))((((((....))))))...................)))))))).(((((....))))).))))))..........------- ( -31.60) >consensus AGUGGCACUCUGAGUGGACUUCCGAGGCGAUGCUCGGACGAUUUAGCAAAGUCGCUGCAGCUCAAACACUAAUUCCAUUCAUUAAAAAACUUUUUAUGUGCCAAUUUUCUUAUUCUUUUU ..((((((..((((((((..((((((......))))))((((((....))))))...................)))))))).(((((....))))).))))))................. (-28.52 = -29.18 + 0.67)

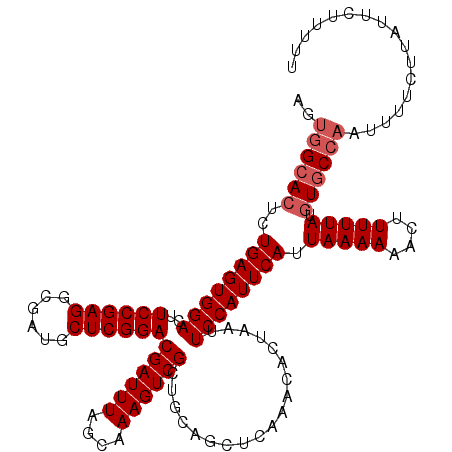

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:44 2006