
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
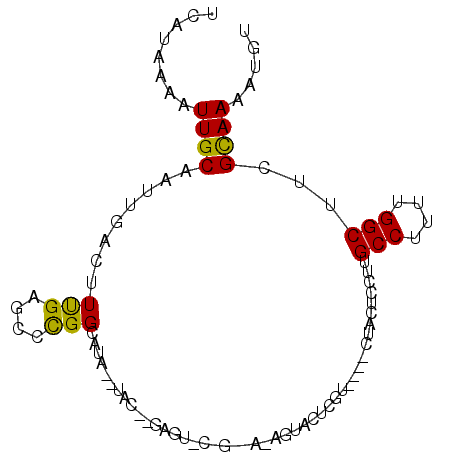
| Location | 13,755,464 – 13,755,559 |
| Length | 95 |
| Max. P | 0.983688 |

| Location | 13,755,464 – 13,755,559 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -6.92 |
| Energy contribution | -6.28 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
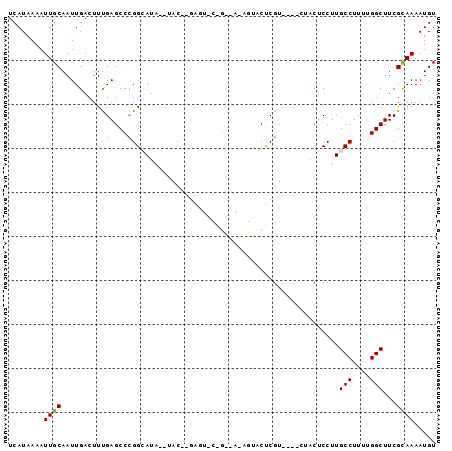
>2L_DroMel_CAF1 13755464 95 + 22407834 UCAUAAAAUUGCAAUUGACUUUGAGCCUGGCAUAUAUAC--GAGUACAGUAAUAGUACUCGUACAUAUUCGCCUUGCCUUUUGGCUUCGCAAAAUGU ........((((................(((((((.(((--((((((.......))))))))).))))..)))..(((....)))...))))..... ( -26.80) >DroSec_CAF1 19090 78 + 1 UCAUAAAAUUGCAAUUGACUUCGAGCCCGGCAUA--UAC--GAG-----------UACUCGU----GUGCUCCUUGCCUUUUGGCUUCGCAAAAUGU ........((((....(....)(((((.((((..--...--(((-----------(((....----))))))..))))....))))).))))..... ( -21.00) >DroSim_CAF1 21201 78 + 1 UCAUAAAAUUGCAAUUGACUUCGAGCCCGGCAUA--UAC--GAG-----------UACUCGU----GUGCUCCUUGCCUUUUGGCUUCGCAAAAUGU ........((((....(....)(((((.((((..--...--(((-----------(((....----))))))..))))....))))).))))..... ( -21.00) >DroEre_CAF1 20979 89 + 1 UCAUAAAAUUGCAAUUGACUUUGAGCCCGGAAUA--UGCAGUAUUCCGGCCAAAG--CCCGC----CGACUCCUUGCCUUUUGGCUUCGUAAAAUGU ........((((......(((((.(((.((((((--(...)))))))))))))))--...((----(((...........)))))...))))..... ( -22.80) >DroYak_CAF1 22218 89 + 1 UCAUAAAAUUGCAAUUGACUUUGAGCCCGGAAUA--CACAGUUUUCCGGCCAAAG--CCCGC----CAACUCCUUGCCUUUUGGCUUCGUAAAAUGU ........((((......(((((.(((.((((.(--(...)).))))))))))))--...((----(((...........)))))...))))..... ( -20.50) >consensus UCAUAAAAUUGCAAUUGACUUUGAGCCCGGCAUA__UAC__GAGU_C_G__A_AGUACUCGU____CUACUCCUUGCCUUUUGGCUUCGCAAAAUGU ........((((........(((....))).............................................(((....)))...))))..... ( -6.92 = -6.28 + -0.64)

| Location | 13,755,464 – 13,755,559 |
|---|---|
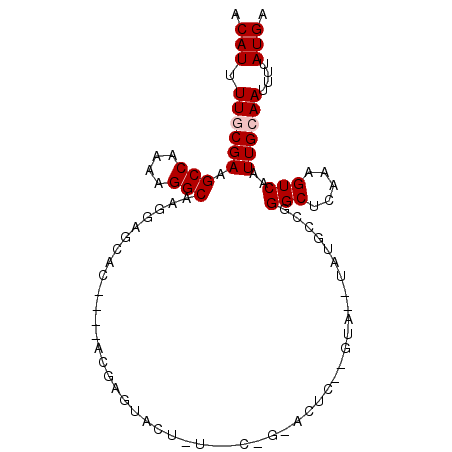
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -10.34 |
| Energy contribution | -10.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
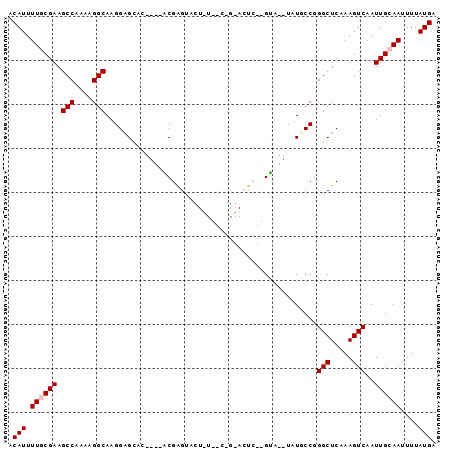
>2L_DroMel_CAF1 13755464 95 - 22407834 ACAUUUUGCGAAGCCAAAAGGCAAGGCGAAUAUGUACGAGUACUAUUACUGUACUC--GUAUAUAUGCCAGGCUCAAAGUCAAUUGCAAUUUUAUGA .(((.((((((.(((....)))..(((..((((((((((((((.......))))))--))))))))))).(((.....)))..))))))....))). ( -36.10) >DroSec_CAF1 19090 78 - 1 ACAUUUUGCGAAGCCAAAAGGCAAGGAGCAC----ACGAGUA-----------CUC--GUA--UAUGCCGGGCUCGAAGUCAAUUGCAAUUUUAUGA .(((.((((((.(((....)))...((((..----.((.(((-----------...--...--..))))).))))........))))))....))). ( -20.10) >DroSim_CAF1 21201 78 - 1 ACAUUUUGCGAAGCCAAAAGGCAAGGAGCAC----ACGAGUA-----------CUC--GUA--UAUGCCGGGCUCGAAGUCAAUUGCAAUUUUAUGA .(((.((((((.(((....)))...((((..----.((.(((-----------...--...--..))))).))))........))))))....))). ( -20.10) >DroEre_CAF1 20979 89 - 1 ACAUUUUACGAAGCCAAAAGGCAAGGAGUCG----GCGGG--CUUUGGCCGGAAUACUGCA--UAUUCCGGGCUCAAAGUCAAUUGCAAUUUUAUGA ........(((..((.........))..)))----((.((--((((((((((((((.....--)))))).))).)))))))....)).......... ( -26.70) >DroYak_CAF1 22218 89 - 1 ACAUUUUACGAAGCCAAAAGGCAAGGAGUUG----GCGGG--CUUUGGCCGGAAAACUGUG--UAUUCCGGGCUCAAAGUCAAUUGCAAUUUUAUGA ............(((....)))..(((((((----...((--((((((((((((.((...)--).)))).))).))))))).....))))))).... ( -25.80) >consensus ACAUUUUGCGAAGCCAAAAGGCAAGGAGCAC____ACGAGUACU_U__C_G_ACUC__GUA__UAUGCCGGGCUCAAAGUCAAUUGCAAUUUUAUGA .(((.((((((.(((....)))................................................(((.....)))..))))))....))). (-10.34 = -10.74 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:39 2006