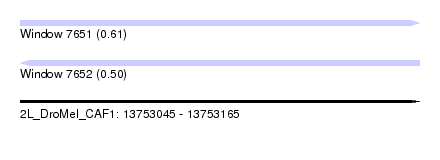
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,753,045 – 13,753,165 |
| Length | 120 |
| Max. P | 0.614059 |

| Location | 13,753,045 – 13,753,165 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -29.01 |
| Energy contribution | -30.57 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
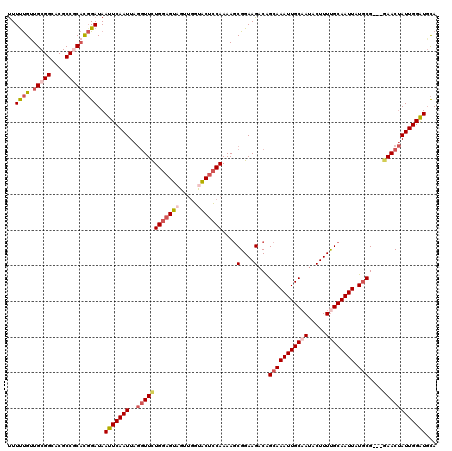
>2L_DroMel_CAF1 13753045 120 + 22407834 UUUUUGUUGCGGCACGCCGCACGGAUAAUUCAAUUAGGUUCUGGAGUAGUUGGUGCUCCAAAAGCGGAAGACAGCAAAUUGCUAUACUUUUGCAAUUAUGCGGAGGAACUAUUGGAUGCA ..(((((.((((....)))))))))..(((((((..((((((((((((.....))))))....(((((((..(((.....)))...)))))))...........)))))))))))))... ( -38.00) >DroSec_CAF1 16706 120 + 1 UUUUUGUUGCGGCACGCCGCACGGAUAAUUCAAUUAGGUUCUGGAGUAGUUGGUGCUCCAAAAGCGGAAGACAGCAAAUUGCAAUACUUUUGCAAUUAUGCGGCGGAACUAUUGGAUGCA ..(((((.((((....)))))))))..(((((((..((((((((((((.....))))))....(((.....).(((((((((((.....)))))))).))).)))))))))))))))... ( -40.40) >DroAna_CAF1 33932 116 + 1 CUUUCGUUGCGGUGCGCCACUUUGAUAAUUCAAUUAAAUUUUGAAGUAGAACUUACAGCAAAA-CGGAAGUCGGCAAAUUGCAAUACUUUCGCAAUUAUGCG---GAACAAUUGGGUCUA .....((((((((..((((((((((....)).......(((((..((((...))))..)))))-..))))).)))..))))))))..(((((((....))))---)))............ ( -26.40) >DroSim_CAF1 18729 120 + 1 UUUUUGUUGCGGCACGCCGCACGGAUAAUUCAAUUAGGUUCUGGAGUAGUUGGUGCUCCAAAAGCGGAAGACAGCAAAUUGCAAUACUUUUGCAAUUAUGCGGAGGAACUAUUGGAUGCA ..(((((.((((....)))))))))..(((((((..((((((((((((.....))))))....((....).).(((((((((((.....)))))))).)))...)))))))))))))... ( -39.90) >DroEre_CAF1 18744 117 + 1 UUUUUGCUGCGGCACGCCACACGGAUAAUUCAAUUAGGUUCUGGAGUAGUUGGCACUCCAAAAGCGGAAGACGGCAAAUUGCAAUACUUUUGCAAUUAUCCG---GAACUAUUGGAUGCA ..((((.((.((....)).))))))..(((((((..(((((((((...((((....(((......)))...)))).((((((((.....)))))))).))))---))))))))))))... ( -35.20) >DroYak_CAF1 19844 117 + 1 UUUCUGUUGCGGCACGCCGCACGGAUAAUUCAAUUAGGUUCUGGAGUAGUUGGCACUCCAAAAGCGGAAGACAGCAAAUUGCAAUACUUUUGCAAUUAUGCG---CAACUAUUGGAUGCA ..(((((.((((....)))))))))..(((((((.((....((((((.(....)))))))...(((.....).(((((((((((.....)))))))).))))---)..)))))))))... ( -38.20) >consensus UUUUUGUUGCGGCACGCCGCACGGAUAAUUCAAUUAGGUUCUGGAGUAGUUGGUACUCCAAAAGCGGAAGACAGCAAAUUGCAAUACUUUUGCAAUUAUGCG___GAACUAUUGGAUGCA ..((((.(((((....)))))))))..(((((((..((((((((((((.....)))))))....(....)...(((((((((((.....)))))))).)))....))))))))))))... (-29.01 = -30.57 + 1.56)

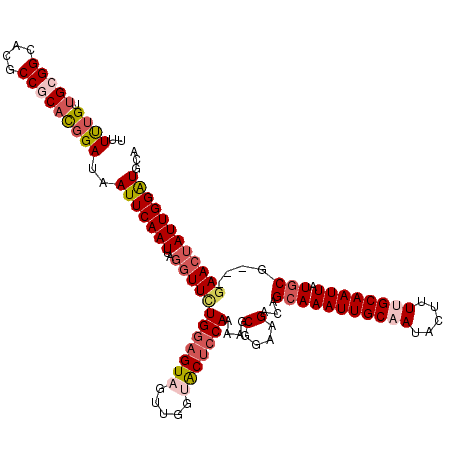
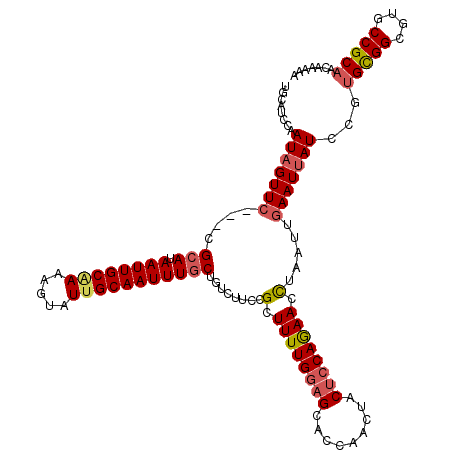
| Location | 13,753,045 – 13,753,165 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -24.29 |
| Energy contribution | -24.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13753045 120 - 22407834 UGCAUCCAAUAGUUCCUCCGCAUAAUUGCAAAAGUAUAGCAAUUUGCUGUCUUCCGCUUUUGGAGCACCAACUACUCCAGAACCUAAUUGAAUUAUCCGUGCGGCGUGCCGCAACAAAAA ........(((((((....(((....)))..(((.(((((.....))))))))..(.((((((((.........)))))))).).....)))))))...(((((....)))))....... ( -28.30) >DroSec_CAF1 16706 120 - 1 UGCAUCCAAUAGUUCCGCCGCAUAAUUGCAAAAGUAUUGCAAUUUGCUGUCUUCCGCUUUUGGAGCACCAACUACUCCAGAACCUAAUUGAAUUAUCCGUGCGGCGUGCCGCAACAAAAA ........(((((((.((.(((.((((((((.....))))))))))).)).....(.((((((((.........)))))))).).....)))))))...(((((....)))))....... ( -30.80) >DroAna_CAF1 33932 116 - 1 UAGACCCAAUUGUUC---CGCAUAAUUGCGAAAGUAUUGCAAUUUGCCGACUUCCG-UUUUGCUGUAAGUUCUACUUCAAAAUUUAAUUGAAUUAUCAAAGUGGCGCACCGCAACGAAAG ......((((((.((---.(((.((((((((.....))))))))))).)).....(-(((((..(((.....)))..)))))).))))))..........((((....))))..(....) ( -22.60) >DroSim_CAF1 18729 120 - 1 UGCAUCCAAUAGUUCCUCCGCAUAAUUGCAAAAGUAUUGCAAUUUGCUGUCUUCCGCUUUUGGAGCACCAACUACUCCAGAACCUAAUUGAAUUAUCCGUGCGGCGUGCCGCAACAAAAA ........(((((((....(((.((((((((.....)))))))))))........(.((((((((.........)))))))).).....)))))))...(((((....)))))....... ( -30.30) >DroEre_CAF1 18744 117 - 1 UGCAUCCAAUAGUUC---CGGAUAAUUGCAAAAGUAUUGCAAUUUGCCGUCUUCCGCUUUUGGAGUGCCAACUACUCCAGAACCUAAUUGAAUUAUCCGUGUGGCGUGCCGCAGCAAAAA (((.....(((((((---(((..((((((((.....))))))))..)))......(.((((((((((.....)))))))))).).....)))))))...(((((....)))))))).... ( -35.40) >DroYak_CAF1 19844 117 - 1 UGCAUCCAAUAGUUG---CGCAUAAUUGCAAAAGUAUUGCAAUUUGCUGUCUUCCGCUUUUGGAGUGCCAACUACUCCAGAACCUAAUUGAAUUAUCCGUGCGGCGUGCCGCAACAGAAA ...........((((---(((((((((((((.....)))))))).(((((.....(.((((((((((.....)))))))))).).....((....))...)))))))).))))))..... ( -32.90) >consensus UGCAUCCAAUAGUUC___CGCAUAAUUGCAAAAGUAUUGCAAUUUGCUGUCUUCCGCUUUUGGAGCACCAACUACUCCAGAACCUAAUUGAAUUAUCCGUGCGGCGUGCCGCAACAAAAA ........(((((((....(((.((((((((.....)))))))))))........(.((((((((.........)))))))).).....)))))))...(((((....)))))....... (-24.29 = -24.85 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:38 2006