| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,751,928 – 13,752,032 |
| Length | 104 |
| Max. P | 0.542408 |

| Location | 13,751,928 – 13,752,032 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -24.88 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
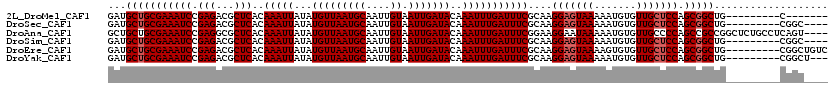
>2L_DroMel_CAF1 13751928 104 - 22407834 GAUGCUGCGAAAUCCGAGACGCUCACAAAUUAUAUGUUAAUGCAAUUGUAAUUGAUACAAAUUUGAUUUCGCAAGGAGUAAAAAUGUGUUGCUCCAGCGGCUG---------C------- ..((((((((((((.(((...)))..(((((...(((((((((....)).)))))))..)))))))))))))..(((((((.......)))))))))))....---------.------- ( -27.30) >DroSec_CAF1 15571 107 - 1 GAUGCUGCGAAAUCCGAGACGCUCACAAAUUAUAUGUUAAUGCAAUUGUAAUUGAUACAAAUUUGAUUUCGCAAGGAGUAAAAAUGUGUUGCUCCAGCGGCUG---------CGGC---- ..((((((((((((.(((...)))..(((((...(((((((((....)).)))))))..)))))))))))))..(((((((.......)))))))))))....---------....---- ( -27.30) >DroAna_CAF1 27206 116 - 1 GCUGCUGCGAAAUCCGAGGCGCUCACAAAUUAUAUGUUAAUGCAAUUGUAAUUGAUACAAAUUUGAUUUCGGAAGGAAUAAAAAUGUGUUGCCCCAGCCGCCGGCUCUGCCUCAGU---- ((....)).......(((((((.((((((((((((((....)))..)))))))........(((.(((((....))))).))).))))..))...((((...))))..)))))...---- ( -28.70) >DroSim_CAF1 17599 107 - 1 GAUGCUGCGAAAUCCGAGACGCUCACAAAUUAUAUGUUAAUGCAAUUGUAAUUGAUACAAAUUUGAUUUCGCAAGGAGUAAAAAUGUGUUGCUCCAGCGGCUG---------CGGC---- ..((((((((((((.(((...)))..(((((...(((((((((....)).)))))))..)))))))))))))..(((((((.......)))))))))))....---------....---- ( -27.30) >DroEre_CAF1 17567 111 - 1 GAUGCUGCGAAAUCCGAGACGCUCACAAAUUAUAUGUUAAUGCAAUUGUAAUUGAUACAAAUUUGAUUUCGCAAGGAGUAAAAGUGUGUUGCUCCAGCGGCUG---------CGGCUGUC .....(((((((((.(((...)))..(((((...(((((((((....)).)))))))..)))))))))))))).(((((((.......))))))).(((((..---------..))))). ( -30.20) >DroYak_CAF1 18675 108 - 1 GAUGCUGCGAAAUCCGAGACGCUCACAAAUUAUAUGUUAAUGCAAUUGUAAUUGAUACAAAUUUGAUUUCGCAAGGAGUAAAAAUGUGUUGCUCCAGCGGCUG---------CGGCU--- ...(((((((((((.(((...)))..(((((...(((((((((....)).)))))))..)))))))))))))..(((((((.......))))))))))(((..---------..)))--- ( -28.50) >consensus GAUGCUGCGAAAUCCGAGACGCUCACAAAUUAUAUGUUAAUGCAAUUGUAAUUGAUACAAAUUUGAUUUCGCAAGGAGUAAAAAUGUGUUGCUCCAGCGGCUG_________CGGC____ ...(((((((((((.(((...)))..(((((...(((((((((....)).)))))))..)))))))))))....(((((((.......))))))).)))))................... (-24.88 = -25.38 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:36 2006